Team:Georgia Tech/Project
From 2012.igem.org
Elsherbini (Talk | contribs) |
Elsherbini (Talk | contribs) (→Design Considerations) |
||
| (29 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Georgia_Tech/Template:stylesheet}} | {{:Team:Georgia_Tech/Template:stylesheet}} | ||
{{:Team:Georgia_Tech/Template:GT-header}} | {{:Team:Georgia_Tech/Template:GT-header}} | ||
| - | |||
| - | |||
| - | |||
==Abstract== | ==Abstract== | ||
| Line 16: | Line 13: | ||
===Motivation=== | ===Motivation=== | ||
| + | Biosensors have a wide range of applications, but for many of those applications there already exists a machine that can do a better job than our engineered bacteria. One of the biggest liabilities in using a biological system is the time associated with getting the desired information. Cells have to metabolize the desired chemical, alter gene expression, and accumulate enough protein in order to produce a measurable output. | ||
| + | |||
| + | [[File:GT_biosensor_example.JPG|thumb|300px|left|A biosensor used in our mentor lab responding to different concentrations of autoinducer]] | ||
| + | |||
| + | In the graph to the left, it takes on the order of 2-3 hours before a LuxR based GFP reporter reaches maximum signal. We designed our system based on the prediction that waiting for dimerization is a lot faster than waiting for transcription, translation, and protein accumulation. | ||
| + | |||
| + | An added benefit to designing our system with TraR to fluoresce independent of TraR's ability to regulate genes is that TraR does not activate transcription in <i>E. coli</i>. [http://www.ncbi.nlm.nih.gov/pubmed/19732344 In a paper] published in 2009 by Qin et. al. they show that TraR is unable to recruit <i> E. coli</i>'s native RNA polymerase. It has been shown in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC126196/ another paper], however, that TraR does fold properly into monomers and will dimerize in the presence of its autoinducer in <i>E. coli</i>. So this system, as we've designed it, will be the first to use TraR in a fluorescence based biosensor in <i>E. coli</i> since it does not require TraR to active transcription but only to fold correctly and dimerize. | ||
===Design Considerations=== | ===Design Considerations=== | ||
| + | |||
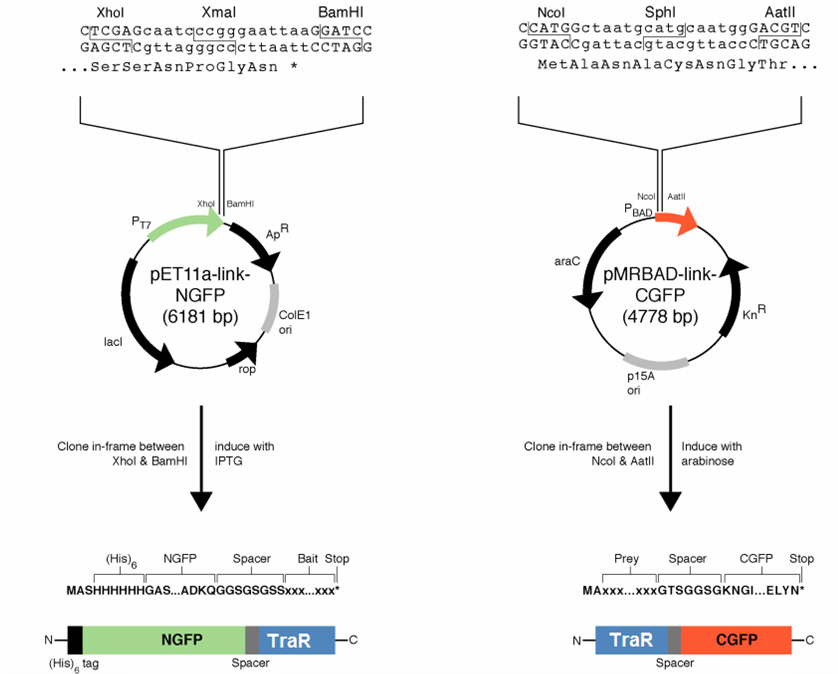
[[File:GT_PET11a_and_pMRBAD.png|right|thumb|250px|Plasmids provided by Dr. Regan from Yale]] | [[File:GT_PET11a_and_pMRBAD.png|right|thumb|250px|Plasmids provided by Dr. Regan from Yale]] | ||
| - | We decided to first make the fusions in the plasmids provided by [http://www.yale.edu/reganlab/ Dr. Lynne Regan] from Yale. By choosing this method, we were able to use the provided controls in testing our part. | + | We decided to first make the fusions in the plasmids provided by [http://www.yale.edu/reganlab/ Dr. Lynne Regan] from Yale. By choosing this method, we were able to use the provided controls in testing our part. [http://www.yale.edu/reganlab/pdfs/WilsonRegan2004NatMethods.pdf Here] is a paper detailing the plasmids. |
The pET11a plasmid is under control of T7 polymerase, so we used Bl21(DE3) <i>E. coli</i> cells which have an IPTG-inducible copy of the polymerase on their chromosome. | The pET11a plasmid is under control of T7 polymerase, so we used Bl21(DE3) <i>E. coli</i> cells which have an IPTG-inducible copy of the polymerase on their chromosome. | ||
| Line 46: | Line 51: | ||
====TraR Fusions==== | ====TraR Fusions==== | ||
| - | [[File:08-20-12_TraR_PCR_products_from_pCF222_and_pCF251_SUCCESS_(Lane_6_elsa_plasmid).jpg|250px|thumb|right|TraR PCR product using pCF222 from Dr. Fuqua]] | + | [[File:08-20-12_TraR_PCR_products_from_pCF222_and_pCF251_SUCCESS_(Lane_6_elsa_plasmid).jpg|250px|thumb|right|TraR PCR product using pCF222 from Dr. Fuqua as template]] |
After trying to PCR traR coding sequence from multiple sources, we finally succeeded using a plasmid generously provided by [http://www.bio.indiana.edu/faculty/directory/profile.php?person=cfuqua Dr. Clay Fuqua] from University of Indiana (pCF222) | After trying to PCR traR coding sequence from multiple sources, we finally succeeded using a plasmid generously provided by [http://www.bio.indiana.edu/faculty/directory/profile.php?person=cfuqua Dr. Clay Fuqua] from University of Indiana (pCF222) | ||
| Line 54: | Line 59: | ||
====Characterization of Fusions==== | ====Characterization of Fusions==== | ||
| + | Diagnostic digests were performed on minipreps from transformed colonies to see if a traR-sized fragment was indeed inserted into the two plasmids. In both plasmids, a ~700bp fragment was observed. These minipreps were sent for sequencing and traR insertion was confirmed and seen to be in frame. | ||
| + | |||
| + | Transformed BL21(DE3) cells carrying both the pET11a-TraR-NGFP and pMRBAD-TraR-CGFP were plated on media containing IPTG, arabinose, and varying amounts of autoinducer, either synthesized, or from sterile supernatants. As of this writing, no conditions produce visible GFP on plates. | ||
| + | |||
| + | Flow cytometry was performed on colonies from the leucine zipper controls, colonies from plates without induction, and colonies grown on plates contiaint 25% supernatants from a strain constitutive for TraI, the enzyme responsible for making TraR's recognized autoinducer. | ||
| + | |||
| + | The leucine zipper colonies were centered around 10<sup>4</sup>, the colonies without induction around 10<sup>1</sup>. The colonies grown on TraI suppernatant supplemented media looked very similar to those without induction, with only a small peak in the range that GFP should produce. | ||
| + | |||
| + | Though this result is not completely positive, some GFP was observed in the TraR fusion colonies. This could mean that with careful optimization of conditions, this system could function as designed. | ||
| + | <gallery> | ||
| + | File:GFP_leucine_zipper_FACS.jpg|leucine zipper control plasmids | ||
| + | File:Negative_FACS.jpg|no induction | ||
| + | File:25%25_supernatants_FACS.jpg|25% TraI supernatants | ||
| + | </gallery> | ||
| + | <br style="clear:both"> | ||
==Submitted Parts== | ==Submitted Parts== | ||
| - | < | + | We successfully cloned 3 of our parts into pSB1c3 before the deadline. |
| + | |||
| + | [http://partsregistry.org/Part:BBa_K916001 TraR-NGFP translational Fusion] | ||
| + | [http://partsregistry.org/Part:BBa_K916002 TraR-CGFP Translational Fusion] | ||
| + | [http://partsregistry.org/Part:BBa_K916003 Leucine Zipper NGFP translational fusion] | ||
| + | |||
| + | However, the NGFP coding sequence contained a Spe1 site. We mistakenly thought that though these parts would be incompatible with some assembly methods, that the parts would still be accepted with proper documentation explaining what assembly methods could and could not be used. We are working with iGEM headquarters to make sure we get compatible DNA submitted to the registry for these parts. | ||
| + | |||
| + | We are in the process of using QuikChange site directed mutagenesis to remove the Spe1 site from BBa_K916001 and BBa_K916003 and will resubmit the parts with compatible sequence as soon as possible. | ||
| + | |||
| + | We sequenced all three parts and saw that all of them were correctly cloned into pSB1c3. All sequencing information can be found on the parts physical DNA page. | ||
| + | |||
| + | <groupparts>iGEM012 Georgia_Tech</groupparts> | ||
| + | |||
| + | ==Characterizing the Las reciever BBa_K091134 from iGEM 2008's Davidson-Missouri Western team== | ||
| + | Our findings have been summarized on the [http://partsregistry.org/Part:BBa_K091134:Experience#Georgia_Tech_2012_iGEM_Team_Review BBa_K091134 experience page]. | ||
| + | ===Background=== | ||
| + | We characterized part BBa_K091134 from the registry which is designed to express GFP in the presence of the autoinducer N-oxo3 dodecanoyl homoserine lactone (''Pseudomonas autoinducer'', PAI). The part is contained in the plasmid psB1AK3 which has markers for ampicillin and kanamycin resistance. It contains the LasR receptor (C0079) from ''Pseudomonas aeruginosa'' and a GFP reporter (E0040). In this part, GFP expression is under the control of the LasR+PAI promoter (R0079). The part should produce LasR which receives the PAI. Binding of LasR to the PAI should be reported by GFP expression. ''lasR'' is reported to be under the control of the PLlac 0-1 promoter (R0011) in the part. The PLlac 0-1 promoter is a hybrid consisting of the lambda phage promoter P(L) with the cI binding sites replaced with lacO1. In nature, LasR bound to the PAI is the activator for the expression of the ''Pseudomonas aeruginosa'' elastase gene (''lasB''). | ||
| + | |||
| + | ===Methods and Results=== | ||
| + | |||
| + | ====Transformation and Selection==== | ||
| + | psB1AK3[BBa_K091134] was transformed into chemically competent (DH5α). Transformed cells were plated on LB agar plates containing either ampicillin, kanamycin, or ampicillin and kanamycin. Colonies were observed on the kan plates and the amp plates but never on the amp and kan plates. Characterization was continued using a colony from the LB/kan plate. | ||
| + | |||
| + | ====Response to Autoinducer==== | ||
| + | |||
| + | In the presence of N-oxo3 dodecanoyl homoserine lactone at various concentrations alone (100 μM, 10 μM, 1μM, 0.1 μM, 0.01 μM, 1 nm, 0.1 nm) and N-oxo3 dodecanoyl homoserine lactone (same concentrations) + IPTG, DH5α/psB1AK3[BBa_K091134] did not produce a detectable fluorescent signal. However,the data are less than clear as our positive control showed no fluorescence either. | ||
| + | |||
| + | ====Sequencing==== | ||
| + | The psB1AK3/BBa_K091134 plasmid was extracted from the transformed cells for sequencing. Standard primers for sequencing/amplifying BioBrick parts were used: VF2 (tgccacctgacgtctaagaa) and VR (attaccgcctttgagtgagc). The sequencing was successful for both primers. EMBOSS Needle was used to align sequencing results to the expected sequence for the part (see attached sequencing data). | ||
| + | |||
| + | The sequence from VF2 from bp 331 to 1099 aligned with the expected sequence from bp 1 to 752 with 92.2% identity. However, it was noted that the region from 332 to 404 bp on the VF2 sequence aligned imperfectly with the expected sequence for that region (1 to 75 bp in the expected sequence for the part). This region is supposed to include the sequence for the PLlac 0-1 promoter (R0011) | ||
| + | |||
| + | However, a comparison between the expected sequence (aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcaca) against the VF2 sequence (agttctggcaggtttggccgcgggttctttttggtacacgaaagc) reveals very little identity (43.1%). This leads us to believe that the promoter region (R0011) for LasR expression is missing and that the part will not function as expected. A BLAST search the VF2 sequence suggests the part does contain the LasR coding sequence (C0079). Directly upstream of the LasR coding sequence is the ribosomal binding site (B0034) which appears to be the correct sequence. However, upstream of the ribosomal binding site, the VF2 sequence BLASTs to the ''P. aeruginosa'' ''lasB'' gene starting at 247 bp. Upstream of ''lasB'' appears to be vector sequence. Nowhere in the sequence is the PLlac 0-1 promoter region (R0011). | ||
| + | |||
| + | The VR sequence (reverse complement) from 1 to 939bp aligns with the expected sequence from 1059 to 2047bp with 97.7% identity. There appears to be no significantly misaligned region. The VR sequence appears to contain the correct sequences for the LasR+PAI promoter (R0079), the ribosomal binding site (B0032), GFP (E0040), and the transcriptional terminator (B0010 and B0012). | ||
| + | |||
| + | ====Raw Sequencing Data==== | ||
| + | |||
| + | =====VF2 Complete Sequence===== | ||
| + | <pre> | ||
| + | NNNNNNNNNNNNCCNATAAAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGC | ||
| + | CAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTG | ||
| + | GGCCTTTCTGCGTTTATATACTAGAGGCCCCTCGCTGAGCGCGTCCCGGAGCTGGGGGCAACCTAGCTGCCACCTGCTTTTCTGCTAGCTATTCCAGCGAAAACATACAG | ||
| + | ATTTCCGGCGAAATCAAGGCTACCTGCCAGTTCTGGCAGGTTTGGCCGCGGGTTCTTTTTGGTACACGAAAGCTACTAGAGAAAGAGGAGAAATACTAGATGGCCTTGGT | ||
| + | TGACGGTTTTCTTGAGCTGGAACGCTCAAGTGGAAAATTGGAGTGGAGCGCCATCCTCCAGAAGATGGCGAGCGACCTTGGATTCTCGAAGATCCTGTTCGGCCTGTTGC | ||
| + | CTAAGGACAGCCAGGACTACGAGAACGCCTTCATCGTCGGCAACTACCCGGCCGCCTGGCGCGAGCATTACGACCGGGCTGGCTACGCGCGGGTCGACCCGACGGTCAGT | ||
| + | CACTGTACCCAGAGCGTACTGCCGATTTTCTGGGAACCGTCCATCTACCAGACGCGAAAGCAGCACGAGTTCTTCGAGGAAGCCTCGGCCGCCGGCCTGGTGTATGGGCT | ||
| + | GACCATGCCGCTGCATGGTGCTCGCGGCGAACTCGGCGCGCTGAGCCTCAGCGTGGAAGCGGAAAACCGGGCCGAGGCCAACCGTTTCATAGAGTCGGTCCTGCCGACCC | ||
| + | TGTGGATGCTCAAGGACTACGCACTGCAAAGCGGTGCCGGACTGGCCTTCGAACATCCGGTCAGCAAACCGGTGGTTCTGACCAGCCGGGANAAGGAAGTGTTGCAGTGG | ||
| + | NGCGCCATCGGCAGACCAGTTGGGAGANATCGGTTATCTGCAACTGCTCGGAAGCCAATGTGAACTTCCATATGGGAAATANTNNNNGGAAGTTCGGNGTGACNNCNNN | ||
| + | </pre> | ||
| + | |||
| + | =====VF2 Alignment with BBa_K091134===== | ||
| + | <pre> | ||
| + | Identity 712/772 (92.2%) | ||
| + | |||
| + | Similarity 712/772 (92.2%) | ||
| + | |||
| + | Gaps 23/772 ( 3.0%) | ||
| + | |||
| + | |||
| + | 1 -ATTTCCGGCGAAATCAAGGCTACCTGCCAGTTCTGGCAGGTTTGGCCGC 49 | ||
| + | |||....|||.| |..||.| ||.|| |||...|| | ||
| + | EMBOSS_001 1 aattgtgagcgga------------taacaat--tgaca---ttgtgagc 33 | ||
| + | |||
| + | EMBOSS_001 50 GGGTTCTTTTTGGTACACGAAAGC-TACTAGAGAAAGAGGAGAAATACTA 98 | ||
| + | ||.| .....|.|||. ||...| ||||||||||||||||||||||||| | ||
| + | EMBOSS_001 34 ggat--aacaagatact-gagcacatactagagaaagaggagaaatacta 80 | ||
| + | |||
| + | EMBOSS_001 99 GATGGCCTTGGTTGACGGTTTTCTTGAGCTGGAACGCTCAAGTGGAAAAT 148 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 81 gatggccttggttgacggttttcttgagctggaacgctcaagtggaaaat 130 | ||
| + | |||
| + | EMBOSS_001 149 TGGAGTGGAGCGCCATCCTCCAGAAGATGGCGAGCGACCTTGGATTCTCG 198 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 131 tggagtggagcgccatcctccagaagatggcgagcgaccttggattctcg 180 | ||
| + | |||
| + | EMBOSS_001 199 AAGATCCTGTTCGGCCTGTTGCCTAAGGACAGCCAGGACTACGAGAACGC 248 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 181 aagatcctgttcggcctgttgcctaaggacagccaggactacgagaacgc 230 | ||
| + | |||
| + | EMBOSS_001 249 CTTCATCGTCGGCAACTACCCGGCCGCCTGGCGCGAGCATTACGACCGGG 298 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 231 cttcatcgtcggcaactacccggccgcctggcgcgagcattacgaccggg 280 | ||
| + | |||
| + | EMBOSS_001 299 CTGGCTACGCGCGGGTCGACCCGACGGTCAGTCACTGTACCCAGAGCGTA 348 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 281 ctggctacgcgcgggtcgacccgacggtcagtcactgtacccagagcgta 330 | ||
| + | |||
| + | EMBOSS_001 349 CTGCCGATTTTCTGGGAACCGTCCATCTACCAGACGCGAAAGCAGCACGA 398 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 331 ctgccgattttctgggaaccgtccatctaccagacgcgaaagcagcacga 380 | ||
| + | |||
| + | EMBOSS_001 399 GTTCTTCGAGGAAGCCTCGGCCGCCGGCCTGGTGTATGGGCTGACCATGC 448 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 381 gttcttcgaggaagcctcggccgccggcctggtgtatgggctgaccatgc 430 | ||
| + | |||
| + | EMBOSS_001 449 CGCTGCATGGTGCTCGCGGCGAACTCGGCGCGCTGAGCCTCAGCGTGGAA 498 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 431 cgctgcatggtgctcgcggcgaactcggcgcgctgagcctcagcgtggaa 480 | ||
| + | |||
| + | EMBOSS_001 499 GCGGAAAACCGGGCCGAGGCCAACCGTTTCATAGAGTCGGTCCTGCCGAC 548 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 481 gcggaaaaccgggccgaggccaaccgtttcatagagtcggtcctgccgac 530 | ||
| + | |||
| + | EMBOSS_001 549 CCTGTGGATGCTCAAGGACTACGCACTGCAAAGCGGTGCCGGACTGGCCT 598 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 531 cctgtggatgctcaaggactacgcactgcaaagcggtgccggactggcct 580 | ||
| + | |||
| + | |||
| + | EMBOSS_001 599 TCGAACATCCGGTCAGCAAACCGGTGGTTCTGACCAGCCGGGANAAGGAA 648 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||.|||||| | ||
| + | EMBOSS_001 581 tcgaacatccggtcagcaaaccggtggttctgaccagccgggagaaggaa 630 | ||
| + | |||
| + | EMBOSS_001 649 GTGTTGCAGTGGNGCGCCATCGGC-AGACCAGTTGGGAGANATCGGTTAT 697 | ||
| + | ||||||||||||.||||||||||| |||||||||||||||.||||||||| | ||
| + | EMBOSS_001 631 gtgttgcagtggtgcgccatcggcaagaccagttgggagatatcggttat 680 | ||
| + | |||
| + | EMBOSS_001 698 CTGCAACTGCTCGGAAGCCAATGTGAACTTCCATATGGGAAATANTNNNN 747 | ||
| + | ||||||||||||||||||||||||||||||||||||||||||||.|.... | ||
| + | EMBOSS_001 681 ctgcaactgctcggaagccaatgtgaacttccatatgggaaatattcggc 730 | ||
| + | |||
| + | EMBOSS_001 748 GGAAGTTCGGNGTGACNNCNNN 769 | ||
| + | ||||||||||.|||||..|... | ||
| + | EMBOSS_001 731 ggaagttcggtgtgacctcccg 752 | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | =====VF2 Misaligned Sequence vs. Expected Sequence===== | ||
| + | <pre> | ||
| + | VF2: agttctggcaggtttggccgcgggttctttttggtacacgaaagc | ||
| + | Expected: aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcaca | ||
| + | </pre> | ||
| + | |||
| + | =====VR Complete Sequence (Reverse Complement)===== | ||
| + | <pre> | ||
| + | NNNTACCTNCCAGTTCNGGCAGGNTTNGNCCNNGNGTTNTTTTTGGTNCNNGAAAGCTACTAGAGTCACNCAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCAC | ||
| + | TGGAGTNGTCCCAATTNTGTTGAATTAGATGGTGATNNAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTT | ||
| + | ATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTT | ||
| + | TTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTG | ||
| + | TTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAA | ||
| + | CAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGT | ||
| + | CCTTTTACCAGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTA | ||
| + | CACATGGCATGGATGAACTATACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGT | ||
| + | GAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACC | ||
| + | CTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGNTCGGTCGTNNNNNNNNNNNNNNNNNNN | ||
| + | </pre> | ||
| + | |||
| + | =====VR Alignment with BBa_K091134===== | ||
| + | |||
| + | <pre> | ||
| + | Identity 919/941 (97.7%) | ||
| + | |||
| + | Similarity 919/941 (97.7%) | ||
| + | |||
| + | Gaps 3/941 ( 0.3%) | ||
| + | |||
| + | |||
| + | EMBOSS_001 1 NNNTACCTNCCAGTTCNGGCAGGNTTNGNCCNNGNGTTNTTTTTGGTNCN 50 | ||
| + | ...|||||.|||||||.|||||| ||.|.||..|.|||.||||||||.|. | ||
| + | EMBOSS_001 1 ggctacctgccagttctggcagg-tttggccgcgggttctttttggtaca 49 | ||
| + | |||
| + | EMBOSS_001 51 NGAAAGCTACTAGAGTCACNCAGGAAAGTACTAGATGCGTAAAGGAGAAG 100 | ||
| + | .||||||||||||||||||.|||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 50 cgaaagctactagagtcacacaggaaagtactagatgcgtaaaggagaag 99 | ||
| + | |||
| + | EMBOSS_001 101 AACTTTTCACTGGAGTNGTCCCAATT-NTGTTGAATTAGATGGTGAT-NN 148 | ||
| + | ||||||||||||||||.||||||||| .||||||||||||||||||| .. | ||
| + | EMBOSS_001 100 aacttttcactggagttgtcccaattcttgttgaattagatggtgatgtt 149 | ||
| + | |||
| + | EMBOSS_001 149 AATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATA 198 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 150 aatgggcacaaattttctgtcagtggagagggtgaaggtgatgcaacata 199 | ||
| + | |||
| + | EMBOSS_001 199 CGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTC 248 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 200 cggaaaacttacccttaaatttatttgcactactggaaaactacctgttc 249 | ||
| + | |||
| + | EMBOSS_001 249 CATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCG 298 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 250 catggccaacacttgtcactactttcggttatggtgttcaatgctttgcg 299 | ||
| + | |||
| + | EMBOSS_001 299 AGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCC 348 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 300 agatacccagatcatatgaaacagcatgactttttcaagagtgccatgcc 349 | ||
| + | |||
| + | EMBOSS_001 349 CGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACT 398 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 350 cgaaggttatgtacaggaaagaactatatttttcaaagatgacgggaact 399 | ||
| + | |||
| + | EMBOSS_001 399 ACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGA 448 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 400 acaagacacgtgctgaagtcaagtttgaaggtgatacccttgttaataga 449 | ||
| + | |||
| + | EMBOSS_001 449 ATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACA 498 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 450 atcgagttaaaaggtattgattttaaagaagatggaaacattcttggaca 499 | ||
| + | |||
| + | EMBOSS_001 499 CAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACA 548 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 500 caaattggaatacaactataactcacacaatgtatacatcatggcagaca 549 | ||
| + | |||
| + | EMBOSS_001 549 AACAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAA 598 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 550 aacaaaagaatggaatcaaagttaacttcaaaattagacacaacattgaa 599 | ||
| + | |||
| + | EMBOSS_001 599 GATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGG 648 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 600 gatggaagcgttcaactagcagaccattatcaacaaaatactccaattgg 649 | ||
| + | |||
| + | EMBOSS_001 649 CGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACACAATCTG 698 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 650 cgatggccctgtccttttaccagacaaccattacctgtccacacaatctg 699 | ||
| + | |||
| + | EMBOSS_001 699 CCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAG 748 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 700 ccctttcgaaagatcccaacgaaaagagagaccacatggtccttcttgag 749 | ||
| + | |||
| + | EMBOSS_001 749 TTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAATA 798 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 750 tttgtaacagctgctgggattacacatggcatggatgaactatacaaata 799 | ||
| + | |||
| + | EMBOSS_001 799 ATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGAC 848 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 800 ataatactagagccaggcatcaaataaaacgaaaggctcagtcgaaagac 849 | ||
| + | |||
| + | EMBOSS_001 849 TGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAG 898 | ||
| + | |||||||||||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 850 tgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagag 899 | ||
| + | |||
| + | EMBOSS_001 899 TCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATA 939 | ||
| + | ||||||||||||||||||||||||||||||||||||||||| | ||
| + | EMBOSS_001 900 tcacactggctcaccttcgggtgggcctttctgcgtttata 940 | ||
| + | </pre> | ||
| + | |||
| + | ===Conclusions=== | ||
| + | |||
| + | From the sequencing data alone, it appears that beyond the expected PLlac 0-1 promoter region, the part is otherwise constructed as reported. However, the lack of the PLlac 0-1 promoter region means that ''lasR'' will not be expressed and there we anticipate that there will be no response (i.e. fluorescence) to the presence of the autoinducer. To fix this part, the ''lasB'' and vector coding sequences upstream of ''lasR'' would need to be cut out and replaced with PLlac 0-1. This is predicted to restore the desired function to the part. | ||
Latest revision as of 04:00, 4 October 2012
Contents
|
Abstract
An intragenic complementation approach to engineer a faster fluorescence biosensor
Our goal is to engineer a novel biosensor with a faster readout than is currently available. Many bacteria produce, secrete, and respond to chemicals called autoinducers to monitor population density and to synchronize gene expression, a process called quorum sensing. In quorum sensing based biosensors, detection of autoinducer activates transcription of a reporter gene, which must then be translated and accumulate to detectable levels, which can take two to four hours. In our system, we will use TraR, a protein used in the quorum sensing response of Agrobacterium tumefaciens, which dimerizes only in the presence of its autoinducer. We have successfully fused traR to sequence for two separate complementary fragments of GFP. Upon addition of autoinducer, we predict that already accumulated TraR-GFP fragment monomers will dimerize, allowing the GFP fragments to interact and fluoresce. This new approach may drastically reduce the time necessary for future biosensors to produce detectable output.
Project Overview
Motivation
Biosensors have a wide range of applications, but for many of those applications there already exists a machine that can do a better job than our engineered bacteria. One of the biggest liabilities in using a biological system is the time associated with getting the desired information. Cells have to metabolize the desired chemical, alter gene expression, and accumulate enough protein in order to produce a measurable output.
In the graph to the left, it takes on the order of 2-3 hours before a LuxR based GFP reporter reaches maximum signal. We designed our system based on the prediction that waiting for dimerization is a lot faster than waiting for transcription, translation, and protein accumulation.
An added benefit to designing our system with TraR to fluoresce independent of TraR's ability to regulate genes is that TraR does not activate transcription in E. coli. [http://www.ncbi.nlm.nih.gov/pubmed/19732344 In a paper] published in 2009 by Qin et. al. they show that TraR is unable to recruit E. coli's native RNA polymerase. It has been shown in [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC126196/ another paper], however, that TraR does fold properly into monomers and will dimerize in the presence of its autoinducer in E. coli. So this system, as we've designed it, will be the first to use TraR in a fluorescence based biosensor in E. coli since it does not require TraR to active transcription but only to fold correctly and dimerize.
Design Considerations
We decided to first make the fusions in the plasmids provided by [http://www.yale.edu/reganlab/ Dr. Lynne Regan] from Yale. By choosing this method, we were able to use the provided controls in testing our part. [http://www.yale.edu/reganlab/pdfs/WilsonRegan2004NatMethods.pdf Here] is a paper detailing the plasmids.
The pET11a plasmid is under control of T7 polymerase, so we used Bl21(DE3) E. coli cells which have an IPTG-inducible copy of the polymerase on their chromosome.
The designed fusion into pET11a was compatible with the TraR sequence, but the pMRBAD fusion was not. The pMRBAD fusion was designed to use NcoI and AatII, but TraR had two AatII sites in the coding sequence. We instead cloned TraR in-between the NcoI and Sph1 sites. This added two amino acids to the linker, one of which was a cysteine. It is unclear whether these additional amino acids will disrupt the ability for TraR and the CGFP fragment to fold independently from one another.
The junction between traR and the C-terminal GFP fragment cloned into the SphI site:
SphI AatII
R K L I A C N G T S G
cgg aaa ctc atc gca tgc aat ggg acg tcg ggt
<-- traR--| |-- linker -->
Results
Leucine Zipper Controls
After co-transforming pET11a-z-NGFP and pMRBAD-z-CGFP into BL21(DE3) cells, we plated on media containing IPTG (0.1mM) and arabinose (0.2% w/v). We incubated at 30° C for 10 hours and left the plates on the bench top for 24 hours. We observed accumulation of GFP only in colonies plated with both IPTG and arabinose.
TraR Fusions
After trying to PCR traR coding sequence from multiple sources, we finally succeeded using a plasmid generously provided by [http://www.bio.indiana.edu/faculty/directory/profile.php?person=cfuqua Dr. Clay Fuqua] from University of Indiana (pCF222)
We successfully cloned traR coding sequence into pET11-a-link-NGFP between the XhoI and BamHI sites, and into pMRBAD-link-CGFP between the NcoI and SphI sites. We confirmed insertion with diagnostic digest and sequencing.
Characterization of Fusions
Diagnostic digests were performed on minipreps from transformed colonies to see if a traR-sized fragment was indeed inserted into the two plasmids. In both plasmids, a ~700bp fragment was observed. These minipreps were sent for sequencing and traR insertion was confirmed and seen to be in frame.
Transformed BL21(DE3) cells carrying both the pET11a-TraR-NGFP and pMRBAD-TraR-CGFP were plated on media containing IPTG, arabinose, and varying amounts of autoinducer, either synthesized, or from sterile supernatants. As of this writing, no conditions produce visible GFP on plates.
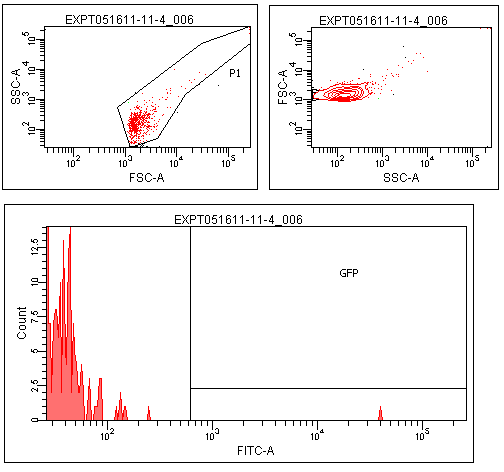
Flow cytometry was performed on colonies from the leucine zipper controls, colonies from plates without induction, and colonies grown on plates contiaint 25% supernatants from a strain constitutive for TraI, the enzyme responsible for making TraR's recognized autoinducer.
The leucine zipper colonies were centered around 104, the colonies without induction around 101. The colonies grown on TraI suppernatant supplemented media looked very similar to those without induction, with only a small peak in the range that GFP should produce.
Though this result is not completely positive, some GFP was observed in the TraR fusion colonies. This could mean that with careful optimization of conditions, this system could function as designed.
Submitted Parts
We successfully cloned 3 of our parts into pSB1c3 before the deadline.
[http://partsregistry.org/Part:BBa_K916001 TraR-NGFP translational Fusion] [http://partsregistry.org/Part:BBa_K916002 TraR-CGFP Translational Fusion] [http://partsregistry.org/Part:BBa_K916003 Leucine Zipper NGFP translational fusion]
However, the NGFP coding sequence contained a Spe1 site. We mistakenly thought that though these parts would be incompatible with some assembly methods, that the parts would still be accepted with proper documentation explaining what assembly methods could and could not be used. We are working with iGEM headquarters to make sure we get compatible DNA submitted to the registry for these parts.
We are in the process of using QuikChange site directed mutagenesis to remove the Spe1 site from BBa_K916001 and BBa_K916003 and will resubmit the parts with compatible sequence as soon as possible.
We sequenced all three parts and saw that all of them were correctly cloned into pSB1c3. All sequencing information can be found on the parts physical DNA page.
<groupparts>iGEM012 Georgia_Tech</groupparts>
Characterizing the Las reciever BBa_K091134 from iGEM 2008's Davidson-Missouri Western team
Our findings have been summarized on the [http://partsregistry.org/Part:BBa_K091134:Experience#Georgia_Tech_2012_iGEM_Team_Review BBa_K091134 experience page].
Background
We characterized part BBa_K091134 from the registry which is designed to express GFP in the presence of the autoinducer N-oxo3 dodecanoyl homoserine lactone (Pseudomonas autoinducer, PAI). The part is contained in the plasmid psB1AK3 which has markers for ampicillin and kanamycin resistance. It contains the LasR receptor (C0079) from Pseudomonas aeruginosa and a GFP reporter (E0040). In this part, GFP expression is under the control of the LasR+PAI promoter (R0079). The part should produce LasR which receives the PAI. Binding of LasR to the PAI should be reported by GFP expression. lasR is reported to be under the control of the PLlac 0-1 promoter (R0011) in the part. The PLlac 0-1 promoter is a hybrid consisting of the lambda phage promoter P(L) with the cI binding sites replaced with lacO1. In nature, LasR bound to the PAI is the activator for the expression of the Pseudomonas aeruginosa elastase gene (lasB).
Methods and Results
Transformation and Selection
psB1AK3[BBa_K091134] was transformed into chemically competent (DH5α). Transformed cells were plated on LB agar plates containing either ampicillin, kanamycin, or ampicillin and kanamycin. Colonies were observed on the kan plates and the amp plates but never on the amp and kan plates. Characterization was continued using a colony from the LB/kan plate.
Response to Autoinducer
In the presence of N-oxo3 dodecanoyl homoserine lactone at various concentrations alone (100 μM, 10 μM, 1μM, 0.1 μM, 0.01 μM, 1 nm, 0.1 nm) and N-oxo3 dodecanoyl homoserine lactone (same concentrations) + IPTG, DH5α/psB1AK3[BBa_K091134] did not produce a detectable fluorescent signal. However,the data are less than clear as our positive control showed no fluorescence either.
Sequencing
The psB1AK3/BBa_K091134 plasmid was extracted from the transformed cells for sequencing. Standard primers for sequencing/amplifying BioBrick parts were used: VF2 (tgccacctgacgtctaagaa) and VR (attaccgcctttgagtgagc). The sequencing was successful for both primers. EMBOSS Needle was used to align sequencing results to the expected sequence for the part (see attached sequencing data).
The sequence from VF2 from bp 331 to 1099 aligned with the expected sequence from bp 1 to 752 with 92.2% identity. However, it was noted that the region from 332 to 404 bp on the VF2 sequence aligned imperfectly with the expected sequence for that region (1 to 75 bp in the expected sequence for the part). This region is supposed to include the sequence for the PLlac 0-1 promoter (R0011)
However, a comparison between the expected sequence (aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcaca) against the VF2 sequence (agttctggcaggtttggccgcgggttctttttggtacacgaaagc) reveals very little identity (43.1%). This leads us to believe that the promoter region (R0011) for LasR expression is missing and that the part will not function as expected. A BLAST search the VF2 sequence suggests the part does contain the LasR coding sequence (C0079). Directly upstream of the LasR coding sequence is the ribosomal binding site (B0034) which appears to be the correct sequence. However, upstream of the ribosomal binding site, the VF2 sequence BLASTs to the P. aeruginosa lasB gene starting at 247 bp. Upstream of lasB appears to be vector sequence. Nowhere in the sequence is the PLlac 0-1 promoter region (R0011).
The VR sequence (reverse complement) from 1 to 939bp aligns with the expected sequence from 1059 to 2047bp with 97.7% identity. There appears to be no significantly misaligned region. The VR sequence appears to contain the correct sequences for the LasR+PAI promoter (R0079), the ribosomal binding site (B0032), GFP (E0040), and the transcriptional terminator (B0010 and B0012).
Raw Sequencing Data
VF2 Complete Sequence
NNNNNNNNNNNNCCNATAAAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGC CAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTG GGCCTTTCTGCGTTTATATACTAGAGGCCCCTCGCTGAGCGCGTCCCGGAGCTGGGGGCAACCTAGCTGCCACCTGCTTTTCTGCTAGCTATTCCAGCGAAAACATACAG ATTTCCGGCGAAATCAAGGCTACCTGCCAGTTCTGGCAGGTTTGGCCGCGGGTTCTTTTTGGTACACGAAAGCTACTAGAGAAAGAGGAGAAATACTAGATGGCCTTGGT TGACGGTTTTCTTGAGCTGGAACGCTCAAGTGGAAAATTGGAGTGGAGCGCCATCCTCCAGAAGATGGCGAGCGACCTTGGATTCTCGAAGATCCTGTTCGGCCTGTTGC CTAAGGACAGCCAGGACTACGAGAACGCCTTCATCGTCGGCAACTACCCGGCCGCCTGGCGCGAGCATTACGACCGGGCTGGCTACGCGCGGGTCGACCCGACGGTCAGT CACTGTACCCAGAGCGTACTGCCGATTTTCTGGGAACCGTCCATCTACCAGACGCGAAAGCAGCACGAGTTCTTCGAGGAAGCCTCGGCCGCCGGCCTGGTGTATGGGCT GACCATGCCGCTGCATGGTGCTCGCGGCGAACTCGGCGCGCTGAGCCTCAGCGTGGAAGCGGAAAACCGGGCCGAGGCCAACCGTTTCATAGAGTCGGTCCTGCCGACCC TGTGGATGCTCAAGGACTACGCACTGCAAAGCGGTGCCGGACTGGCCTTCGAACATCCGGTCAGCAAACCGGTGGTTCTGACCAGCCGGGANAAGGAAGTGTTGCAGTGG NGCGCCATCGGCAGACCAGTTGGGAGANATCGGTTATCTGCAACTGCTCGGAAGCCAATGTGAACTTCCATATGGGAAATANTNNNNGGAAGTTCGGNGTGACNNCNNN
VF2 Alignment with BBa_K091134
Identity 712/772 (92.2%)
Similarity 712/772 (92.2%)
Gaps 23/772 ( 3.0%)
1 -ATTTCCGGCGAAATCAAGGCTACCTGCCAGTTCTGGCAGGTTTGGCCGC 49
|||....|||.| |..||.| ||.|| |||...||
EMBOSS_001 1 aattgtgagcgga------------taacaat--tgaca---ttgtgagc 33
EMBOSS_001 50 GGGTTCTTTTTGGTACACGAAAGC-TACTAGAGAAAGAGGAGAAATACTA 98
||.| .....|.|||. ||...| |||||||||||||||||||||||||
EMBOSS_001 34 ggat--aacaagatact-gagcacatactagagaaagaggagaaatacta 80
EMBOSS_001 99 GATGGCCTTGGTTGACGGTTTTCTTGAGCTGGAACGCTCAAGTGGAAAAT 148
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 81 gatggccttggttgacggttttcttgagctggaacgctcaagtggaaaat 130
EMBOSS_001 149 TGGAGTGGAGCGCCATCCTCCAGAAGATGGCGAGCGACCTTGGATTCTCG 198
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 131 tggagtggagcgccatcctccagaagatggcgagcgaccttggattctcg 180
EMBOSS_001 199 AAGATCCTGTTCGGCCTGTTGCCTAAGGACAGCCAGGACTACGAGAACGC 248
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 181 aagatcctgttcggcctgttgcctaaggacagccaggactacgagaacgc 230
EMBOSS_001 249 CTTCATCGTCGGCAACTACCCGGCCGCCTGGCGCGAGCATTACGACCGGG 298
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 231 cttcatcgtcggcaactacccggccgcctggcgcgagcattacgaccggg 280
EMBOSS_001 299 CTGGCTACGCGCGGGTCGACCCGACGGTCAGTCACTGTACCCAGAGCGTA 348
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 281 ctggctacgcgcgggtcgacccgacggtcagtcactgtacccagagcgta 330
EMBOSS_001 349 CTGCCGATTTTCTGGGAACCGTCCATCTACCAGACGCGAAAGCAGCACGA 398
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 331 ctgccgattttctgggaaccgtccatctaccagacgcgaaagcagcacga 380
EMBOSS_001 399 GTTCTTCGAGGAAGCCTCGGCCGCCGGCCTGGTGTATGGGCTGACCATGC 448
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 381 gttcttcgaggaagcctcggccgccggcctggtgtatgggctgaccatgc 430
EMBOSS_001 449 CGCTGCATGGTGCTCGCGGCGAACTCGGCGCGCTGAGCCTCAGCGTGGAA 498
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 431 cgctgcatggtgctcgcggcgaactcggcgcgctgagcctcagcgtggaa 480
EMBOSS_001 499 GCGGAAAACCGGGCCGAGGCCAACCGTTTCATAGAGTCGGTCCTGCCGAC 548
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 481 gcggaaaaccgggccgaggccaaccgtttcatagagtcggtcctgccgac 530
EMBOSS_001 549 CCTGTGGATGCTCAAGGACTACGCACTGCAAAGCGGTGCCGGACTGGCCT 598
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 531 cctgtggatgctcaaggactacgcactgcaaagcggtgccggactggcct 580
EMBOSS_001 599 TCGAACATCCGGTCAGCAAACCGGTGGTTCTGACCAGCCGGGANAAGGAA 648
|||||||||||||||||||||||||||||||||||||||||||.||||||
EMBOSS_001 581 tcgaacatccggtcagcaaaccggtggttctgaccagccgggagaaggaa 630
EMBOSS_001 649 GTGTTGCAGTGGNGCGCCATCGGC-AGACCAGTTGGGAGANATCGGTTAT 697
||||||||||||.||||||||||| |||||||||||||||.|||||||||
EMBOSS_001 631 gtgttgcagtggtgcgccatcggcaagaccagttgggagatatcggttat 680
EMBOSS_001 698 CTGCAACTGCTCGGAAGCCAATGTGAACTTCCATATGGGAAATANTNNNN 747
||||||||||||||||||||||||||||||||||||||||||||.|....
EMBOSS_001 681 ctgcaactgctcggaagccaatgtgaacttccatatgggaaatattcggc 730
EMBOSS_001 748 GGAAGTTCGGNGTGACNNCNNN 769
||||||||||.|||||..|...
EMBOSS_001 731 ggaagttcggtgtgacctcccg 752
VF2 Misaligned Sequence vs. Expected Sequence
VF2: agttctggcaggtttggccgcgggttctttttggtacacgaaagc
Expected: aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcaca
VR Complete Sequence (Reverse Complement)
NNNTACCTNCCAGTTCNGGCAGGNTTNGNCCNNGNGTTNTTTTTGGTNCNNGAAAGCTACTAGAGTCACNCAGGAAAGTACTAGATGCGTAAAGGAGAAGAACTTTTCAC TGGAGTNGTCCCAATTNTGTTGAATTAGATGGTGATNNAATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATACGGAAAACTTACCCTTAAATTT ATTTGCACTACTGGAAAACTACCTGTTCCATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCGAGATACCCAGATCATATGAAACAGCATGACTT TTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACTACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTG TTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACACAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACAAA CAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAAGATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGT CCTTTTACCAGACAACCATTACCTGTCCACACAATCTGCCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAGTTTGTAACAGCTGCTGGGATTA CACATGGCATGGATGAACTATACAAATAATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGT GAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATATACTAGTAGCGGCCGCTGCAGTCCGGCAAAAAAGGGCAAGGTGTCACCACC CTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCGNTCGGTCGTNNNNNNNNNNNNNNNNNNN
VR Alignment with BBa_K091134
Identity 919/941 (97.7%)
Similarity 919/941 (97.7%)
Gaps 3/941 ( 0.3%)
EMBOSS_001 1 NNNTACCTNCCAGTTCNGGCAGGNTTNGNCCNNGNGTTNTTTTTGGTNCN 50
...|||||.|||||||.|||||| ||.|.||..|.|||.||||||||.|.
EMBOSS_001 1 ggctacctgccagttctggcagg-tttggccgcgggttctttttggtaca 49
EMBOSS_001 51 NGAAAGCTACTAGAGTCACNCAGGAAAGTACTAGATGCGTAAAGGAGAAG 100
.||||||||||||||||||.||||||||||||||||||||||||||||||
EMBOSS_001 50 cgaaagctactagagtcacacaggaaagtactagatgcgtaaaggagaag 99
EMBOSS_001 101 AACTTTTCACTGGAGTNGTCCCAATT-NTGTTGAATTAGATGGTGAT-NN 148
||||||||||||||||.||||||||| .||||||||||||||||||| ..
EMBOSS_001 100 aacttttcactggagttgtcccaattcttgttgaattagatggtgatgtt 149
EMBOSS_001 149 AATGGGCACAAATTTTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACATA 198
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 150 aatgggcacaaattttctgtcagtggagagggtgaaggtgatgcaacata 199
EMBOSS_001 199 CGGAAAACTTACCCTTAAATTTATTTGCACTACTGGAAAACTACCTGTTC 248
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 200 cggaaaacttacccttaaatttatttgcactactggaaaactacctgttc 249
EMBOSS_001 249 CATGGCCAACACTTGTCACTACTTTCGGTTATGGTGTTCAATGCTTTGCG 298
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 250 catggccaacacttgtcactactttcggttatggtgttcaatgctttgcg 299
EMBOSS_001 299 AGATACCCAGATCATATGAAACAGCATGACTTTTTCAAGAGTGCCATGCC 348
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 300 agatacccagatcatatgaaacagcatgactttttcaagagtgccatgcc 349
EMBOSS_001 349 CGAAGGTTATGTACAGGAAAGAACTATATTTTTCAAAGATGACGGGAACT 398
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 350 cgaaggttatgtacaggaaagaactatatttttcaaagatgacgggaact 399
EMBOSS_001 399 ACAAGACACGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGA 448
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 400 acaagacacgtgctgaagtcaagtttgaaggtgatacccttgttaataga 449
EMBOSS_001 449 ATCGAGTTAAAAGGTATTGATTTTAAAGAAGATGGAAACATTCTTGGACA 498
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 450 atcgagttaaaaggtattgattttaaagaagatggaaacattcttggaca 499
EMBOSS_001 499 CAAATTGGAATACAACTATAACTCACACAATGTATACATCATGGCAGACA 548
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 500 caaattggaatacaactataactcacacaatgtatacatcatggcagaca 549
EMBOSS_001 549 AACAAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTGAA 598
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 550 aacaaaagaatggaatcaaagttaacttcaaaattagacacaacattgaa 599
EMBOSS_001 599 GATGGAAGCGTTCAACTAGCAGACCATTATCAACAAAATACTCCAATTGG 648
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 600 gatggaagcgttcaactagcagaccattatcaacaaaatactccaattgg 649
EMBOSS_001 649 CGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTCCACACAATCTG 698
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 650 cgatggccctgtccttttaccagacaaccattacctgtccacacaatctg 699
EMBOSS_001 699 CCCTTTCGAAAGATCCCAACGAAAAGAGAGACCACATGGTCCTTCTTGAG 748
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 700 ccctttcgaaagatcccaacgaaaagagagaccacatggtccttcttgag 749
EMBOSS_001 749 TTTGTAACAGCTGCTGGGATTACACATGGCATGGATGAACTATACAAATA 798
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 750 tttgtaacagctgctgggattacacatggcatggatgaactatacaaata 799
EMBOSS_001 799 ATAATACTAGAGCCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGAC 848
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 800 ataatactagagccaggcatcaaataaaacgaaaggctcagtcgaaagac 849
EMBOSS_001 849 TGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAG 898
||||||||||||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 850 tgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagag 899
EMBOSS_001 899 TCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATA 939
|||||||||||||||||||||||||||||||||||||||||
EMBOSS_001 900 tcacactggctcaccttcgggtgggcctttctgcgtttata 940
Conclusions
From the sequencing data alone, it appears that beyond the expected PLlac 0-1 promoter region, the part is otherwise constructed as reported. However, the lack of the PLlac 0-1 promoter region means that lasR will not be expressed and there we anticipate that there will be no response (i.e. fluorescence) to the presence of the autoinducer. To fix this part, the lasB and vector coding sequences upstream of lasR would need to be cut out and replaced with PLlac 0-1. This is predicted to restore the desired function to the part.
 "
"