Team:Georgia Tech/sandbox/Project/
From 2012.igem.org
Elsherbini (Talk | contribs) |
Elsherbini (Talk | contribs) |
||
| (39 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Georgia_Tech/Template:stylesheet}} | {{:Team:Georgia_Tech/Template:stylesheet}} | ||
{{:Team:Georgia_Tech/Template:GT-header}} | {{:Team:Georgia_Tech/Template:GT-header}} | ||
| + | |||
| + | [https://igem.org/2012_Judging_Form?id=916 Judging Form] | ||
| + | |||
| + | |||
| + | ==Abstract== | ||
| + | |||
===An intragenic complementation approach to engineer a faster fluorescence biosensor=== | ===An intragenic complementation approach to engineer a faster fluorescence biosensor=== | ||
| - | Our goal is to engineer a novel biosensor with a faster readout than is currently available. Many bacteria produce, secrete, and respond to chemicals called autoinducers to monitor population density and to synchronize gene expression, a process called quorum sensing. In quorum sensing based biosensors, detection of autoinducer activates transcription of a reporter gene, which must then be translated and accumulate to detectable levels, which can take two to four hours. In our system, we will use TraR, a protein used in the quorum sensing response of Agrobacterium tumefaciens, which dimerizes only in the presence of its autoinducer. We have successfully fused traR to sequence for two separate complementary fragments of GFP. Upon addition of autoinducer, we predict that already accumulated TraR-GFP fragment monomers will dimerize, allowing the GFP fragments to interact and fluoresce. This new approach may drastically reduce the time necessary for future biosensors to produce detectable output. | + | Our goal is to engineer a novel biosensor with a faster readout than is currently available. Many bacteria produce, secrete, and respond to chemicals called autoinducers to monitor population density and to synchronize gene expression, a process called quorum sensing. In quorum sensing based biosensors, detection of autoinducer activates transcription of a reporter gene, which must then be translated and accumulate to detectable levels, which can take two to four hours. In our system, we will use TraR, a protein used in the quorum sensing response of <i>Agrobacterium tumefaciens</i>, which dimerizes only in the presence of its autoinducer. We have successfully fused traR to sequence for two separate complementary fragments of GFP. Upon addition of autoinducer, we predict that already accumulated TraR-GFP fragment monomers will dimerize, allowing the GFP fragments to interact and fluoresce. This new approach may drastically reduce the time necessary for future biosensors to produce detectable output. |
| + | |||
| + | |||
| + | ==Project Overview== | ||
| + | |||
| + | ===Motivation=== | ||
| + | |||
| + | |||
| + | ===Design Considerations=== | ||
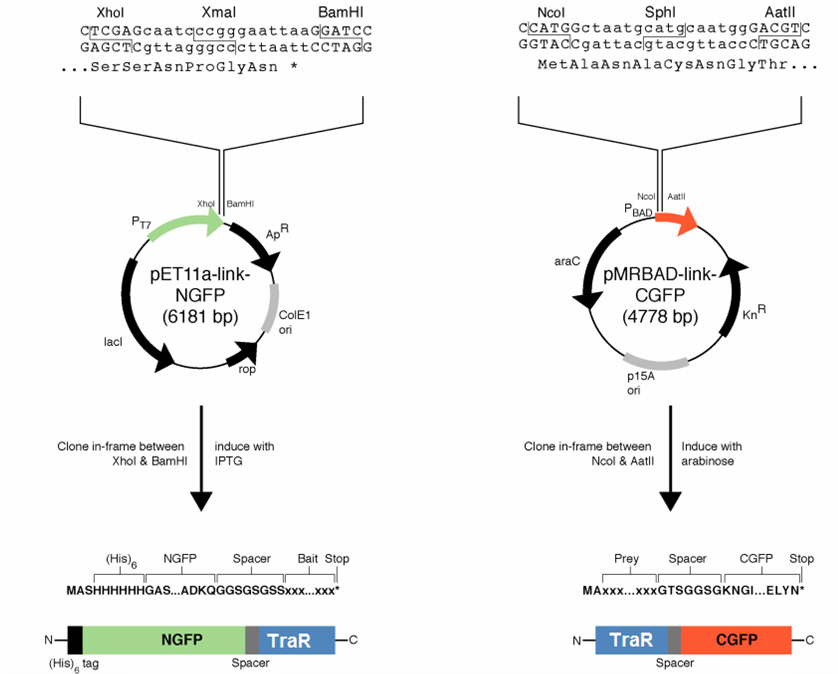
| + | [[File:GT_PET11a_and_pMRBAD.png|right|thumb|250px|Plasmids provided by Dr. Regan from Yale]] | ||
| + | |||
| + | We decided to first make the fusions in the plasmids provided by [http://www.yale.edu/reganlab/ Dr. Lynne Regan] from Yale. By choosing this method, we were able to use the provided controls in testing our part. | ||
| + | |||
| + | The pET11a plasmid is under control of T7 polymerase, so we used Bl21(DE3) <i>E. coli</i> cells which have an IPTG-inducible copy of the polymerase on their chromosome. | ||
| + | |||
| + | The designed fusion into pET11a was compatible with the TraR sequence, but the pMRBAD fusion was not. The pMRBAD fusion was designed to use NcoI and AatII, but TraR had two AatII sites in the coding sequence. We instead cloned TraR in-between the NcoI and Sph1 sites. This added two amino acids to the linker, one of which was a cysteine. It is unclear whether these additional amino acids will disrupt the ability for TraR and the CGFP fragment to fold independently from one another. | ||
| + | |||
| + | <br style="clear:both"> | ||
| + | The junction between traR and the C-terminal GFP fragment cloned into the SphI site: | ||
| + | <pre> | ||
| + | SphI AatII | ||
| + | R K L I A C N G T S G | ||
| + | cgg aaa ctc atc gca tgc aat ggg acg tcg ggt | ||
| + | <-- traR--| |-- linker --> </pre> | ||
| + | |||
| + | ===Results=== | ||
| + | |||
| + | |||
| + | |||
| + | ====Leucine Zipper Controls==== | ||
| + | |||
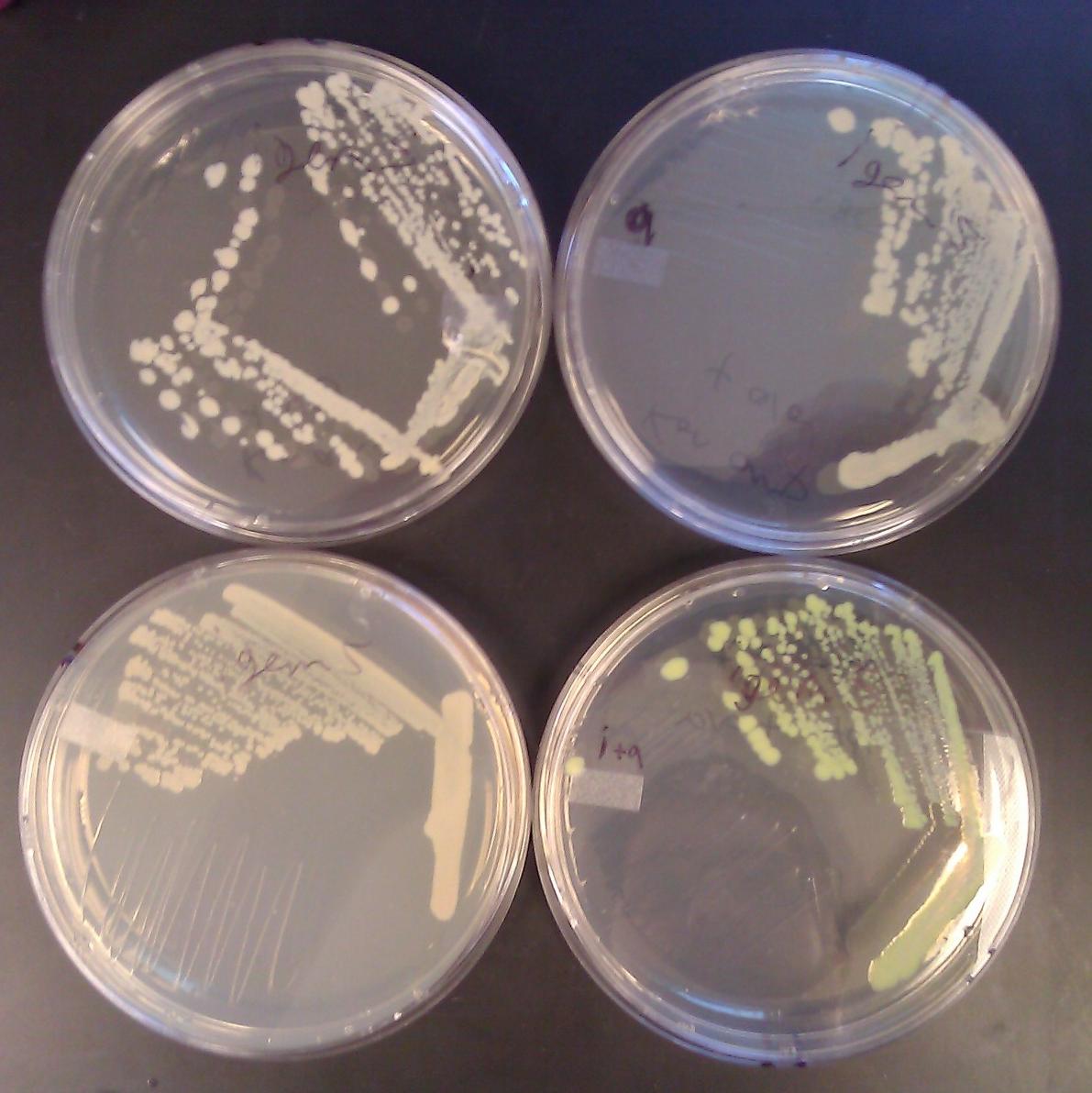
| + | [[File:GT_leucine_zipper_GFP_plates.JPG|250px|thumb|right|Leucine zipper controls glow only upon addition of both arabinose and IPTG]] | ||
| + | |||
| + | After co-transforming pET11a-z-NGFP and pMRBAD-z-CGFP into BL21(DE3) cells, we plated on media containing IPTG (0.1mM) and arabinose (0.2% w/v). We incubated at 30° C for 10 hours and left the plates on the bench top for 24 hours. We observed accumulation of GFP only in colonies plated with both IPTG and arabinose. | ||
| + | |||
| + | ====TraR Fusions==== | ||
| + | |||
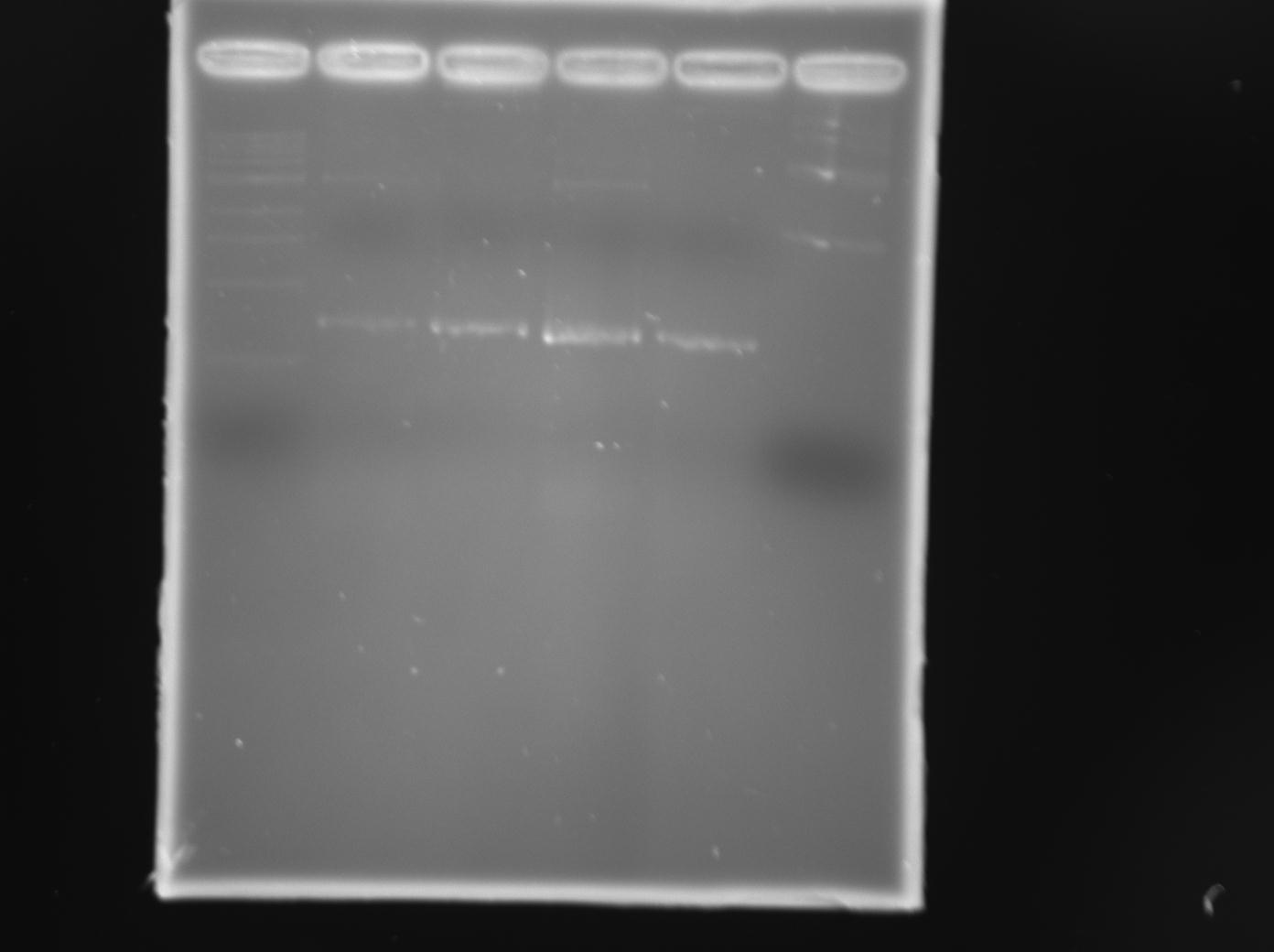
| + | [[File:08-20-12_TraR_PCR_products_from_pCF222_and_pCF251_SUCCESS_(Lane_6_elsa_plasmid).jpg|250px|thumb|right|TraR PCR product using pCF222 from Dr. Fuqua]] | ||
| + | |||
| + | After trying to PCR traR coding sequence from multiple sources, we finally succeeded using a plasmid generously provided by [http://www.bio.indiana.edu/faculty/directory/profile.php?person=cfuqua Dr. Clay Fuqua] from University of Indiana (pCF222) | ||
| + | |||
| + | We successfully cloned traR coding sequence into pET11-a-link-NGFP between the XhoI and BamHI sites, and into pMRBAD-link-CGFP between the NcoI and SphI sites. We confirmed insertion with diagnostic digest and sequencing. | ||
| + | |||
| + | ====Characterization of Fusions==== | ||
| + | |||
| + | |||
| + | ==Submitted Parts== | ||
| + | <br style="clear:both"><groupparts>iGEM012 Georgia_Tech</groupparts> | ||
Latest revision as of 01:22, 3 October 2012
Contents |
Abstract
An intragenic complementation approach to engineer a faster fluorescence biosensor
Our goal is to engineer a novel biosensor with a faster readout than is currently available. Many bacteria produce, secrete, and respond to chemicals called autoinducers to monitor population density and to synchronize gene expression, a process called quorum sensing. In quorum sensing based biosensors, detection of autoinducer activates transcription of a reporter gene, which must then be translated and accumulate to detectable levels, which can take two to four hours. In our system, we will use TraR, a protein used in the quorum sensing response of Agrobacterium tumefaciens, which dimerizes only in the presence of its autoinducer. We have successfully fused traR to sequence for two separate complementary fragments of GFP. Upon addition of autoinducer, we predict that already accumulated TraR-GFP fragment monomers will dimerize, allowing the GFP fragments to interact and fluoresce. This new approach may drastically reduce the time necessary for future biosensors to produce detectable output.
Project Overview
Motivation
Design Considerations
We decided to first make the fusions in the plasmids provided by [http://www.yale.edu/reganlab/ Dr. Lynne Regan] from Yale. By choosing this method, we were able to use the provided controls in testing our part.
The pET11a plasmid is under control of T7 polymerase, so we used Bl21(DE3) E. coli cells which have an IPTG-inducible copy of the polymerase on their chromosome.
The designed fusion into pET11a was compatible with the TraR sequence, but the pMRBAD fusion was not. The pMRBAD fusion was designed to use NcoI and AatII, but TraR had two AatII sites in the coding sequence. We instead cloned TraR in-between the NcoI and Sph1 sites. This added two amino acids to the linker, one of which was a cysteine. It is unclear whether these additional amino acids will disrupt the ability for TraR and the CGFP fragment to fold independently from one another.
The junction between traR and the C-terminal GFP fragment cloned into the SphI site:
SphI AatII
R K L I A C N G T S G
cgg aaa ctc atc gca tgc aat ggg acg tcg ggt
<-- traR--| |-- linker -->
Results
Leucine Zipper Controls
After co-transforming pET11a-z-NGFP and pMRBAD-z-CGFP into BL21(DE3) cells, we plated on media containing IPTG (0.1mM) and arabinose (0.2% w/v). We incubated at 30° C for 10 hours and left the plates on the bench top for 24 hours. We observed accumulation of GFP only in colonies plated with both IPTG and arabinose.
TraR Fusions
After trying to PCR traR coding sequence from multiple sources, we finally succeeded using a plasmid generously provided by [http://www.bio.indiana.edu/faculty/directory/profile.php?person=cfuqua Dr. Clay Fuqua] from University of Indiana (pCF222)
We successfully cloned traR coding sequence into pET11-a-link-NGFP between the XhoI and BamHI sites, and into pMRBAD-link-CGFP between the NcoI and SphI sites. We confirmed insertion with diagnostic digest and sequencing.
Characterization of Fusions
Submitted Parts
<groupparts>iGEM012 Georgia_Tech</groupparts>
 "
"