Team:Gaston Day School/Project
From 2012.igem.org
| (19 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:Gaston_Day_School/Templates/Top}} | {{Template:Team:Gaston_Day_School/Templates/Top}} | ||
| - | + | <html> | |
| + | <br> | ||
| + | <h1><center>Overall Project</h1> | ||
| + | </html> | ||
{|align="justify" | {|align="justify" | ||
| | | | ||
| Line 11: | Line 14: | ||
| | | | ||
|} | |} | ||
| - | + | <html> | |
| - | + | <br> | |
| - | + | <h1><center>Project Details</h1> | |
| - | We built the GFP Cadmium and Arsenic Sensors using NEB's BioBrick Construction Kit. We digested the promoter responsive to | + | </html> |
| + | <div style="text-align: left;"> | ||
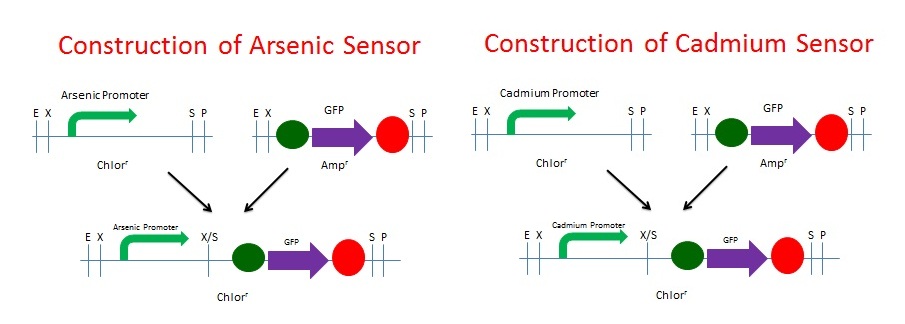
| + | We built the GFP Cadmium and Arsenic Sensors using NEB's BioBrick Construction Kit. We digested the promoter responsive to Cadmium (BBa_K174015) with EcoRI and SpeI and digested the GFP plasmid (BBa_I13401) with XbaI and PstI. The same process was used for Arsenic (BBa_J33201). The promoters were Chloramphenicol resistant and the GFP was Ampicillin resistant. These digested fragments were mixed and ligated to the provided, linearized pSB1C3 plasmid. The ligation mix was grown under Chloramphenicol selection. The resulting colonies were tested for responsive GFP production from the addition of Cadmium. We chose Cadmium because of our BSL. We are not allowed to have Arsenic on campus for testing since it is a known poison. Schematics of the 2 processes are shown below. | ||
<div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:Constructsarscad.jpg|900px]] | <div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:Constructsarscad.jpg|900px]] | ||
| Line 21: | Line 26: | ||
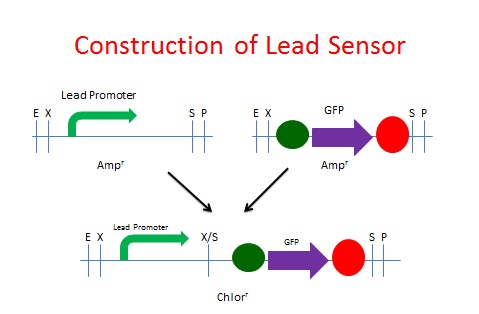
| - | + | The same process was used to create our Lead Sensor. A lead responsive promoter (BBa_I721001) was paired with GFP (BBa_I13401). However, the [http://partsregistry.org/Part:BBa_I721001 lead responsive promoter] was not known to function, according to the [http://openwetware.org/wiki/IGEM:Brown/2007 2007 Brown iGEM team]. The schematic process is shown below. | |
| - | + | ||
| - | + | <div class="center" style="width:auto; margin-left:auto; margin-right:auto;">[[Image:leadconst.jpg|500px]] | |
| - | + | </div> | |
| + | <html> | ||
| + | <br> | ||
| + | <h1><center>The Experiments</h1> | ||
| + | </html> | ||
| + | <div style="text-align: left;"> | ||
| + | We tested out detectors (protocol can be found [https://static.igem.org/mediawiki/2012/d/da/2012GDS8-20.23.24.pdf here]) and measured the sensitivity using a spectrophotometer. We measured in Flu./OD which is the amount of fluorescence per OD (at 600nm). At first, we encountered many problems. We noticed that all of our dilutions, including the control were approximately the same. We realized that LB (Luria Broth, our growth medium) fluoresces naturally; hence, it masked the effects of the GFP. To solve this problem, we tried to run our tests using M9 Salts as a minimal growth medium. However, our bacteria were unable to grow in the medium. After this, our solution was to grow in LB, spin down the bacteria, then resuspend in PBS (phosphate buffer solution). Lastly, our spectrophotometer still continued to show the same fluorescence reading for most samples except the control. We then realized that the readings had maxed. Serial dilutions were used to determine accurate measurements. | ||
| + | |||
| + | <html> | ||
| + | <br> | ||
| + | <h1><center>Results</h1> | ||
| + | </html> | ||
| + | <div style="text-align: left;"> | ||
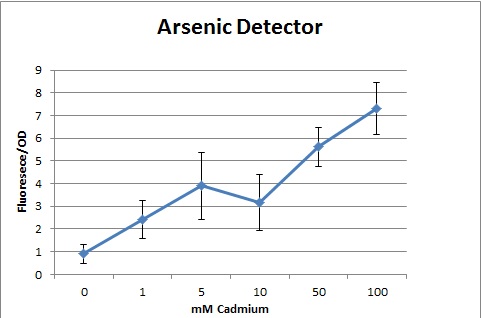
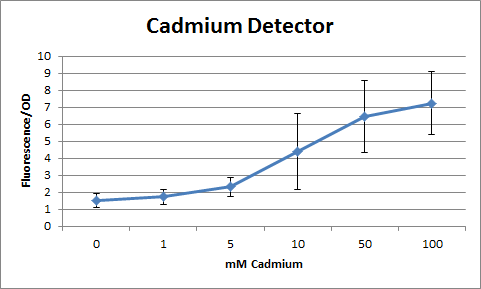
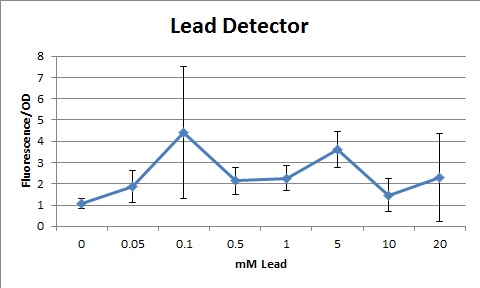
Overall, our tests show that all 3 of our detectors worked properly, even the lead detector with the "non-functional" lead promoter. | Overall, our tests show that all 3 of our detectors worked properly, even the lead detector with the "non-functional" lead promoter. | ||
| + | |||
| + | The graphs shown below represent our results (in Fluorescence per OD600) for the three detectors. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | [[File:Arsgraph.jpg|center|400px|caption]] | ||
| + | <br/> | ||
| + | [[File:Cadgraphuse.jpg|center|400px|caption]] | ||
| + | <br/> | ||
| + | [[File:Leadgraph.jpg|center|400px|caption]] | ||
Latest revision as of 20:55, 3 October 2012
Overall Project
|
Heavy metal contaminants in water pose serious health problems; the lungs, liver, kidneys, blood, digestive system, and the nervous system are all affected by contamination. The Agency for Toxic Substances and Disease registry released a Priority List of Hazardous Substances (ASTDR). Heavy metals accounted for almost half of the top 10 substances; therefore, we have constructed a set of sensors that detects heavy metal contaminants in water. Our sensors provide an inexpensive, simple, and visual test for Arsenic (the number one substance from ASTDR’s list), Lead (number two), and Cadmium (number seven). One sensor paired a promoter responsive to both Cadmium and Arsenic with GFP as a reporter. Another was created for the detection of Lead and a third sensor was specific to cadmium. Use of the detector could potentially save lives around the world through early detection of the contamination. The kit will include all necessary components for running the test and then decontaminating the resulting growth to prevent release of the engineered bacteria into the environment. We will include a very simple mechanism for killing or denaturing the bacteria in our detector kit – bleach. Bleach is highly effective at killing bacteria and is readily available to the average person. |
Project Details
We built the GFP Cadmium and Arsenic Sensors using NEB's BioBrick Construction Kit. We digested the promoter responsive to Cadmium (BBa_K174015) with EcoRI and SpeI and digested the GFP plasmid (BBa_I13401) with XbaI and PstI. The same process was used for Arsenic (BBa_J33201). The promoters were Chloramphenicol resistant and the GFP was Ampicillin resistant. These digested fragments were mixed and ligated to the provided, linearized pSB1C3 plasmid. The ligation mix was grown under Chloramphenicol selection. The resulting colonies were tested for responsive GFP production from the addition of Cadmium. We chose Cadmium because of our BSL. We are not allowed to have Arsenic on campus for testing since it is a known poison. Schematics of the 2 processes are shown below.
The same process was used to create our Lead Sensor. A lead responsive promoter (BBa_I721001) was paired with GFP (BBa_I13401). However, the [http://partsregistry.org/Part:BBa_I721001 lead responsive promoter] was not known to function, according to the [http://openwetware.org/wiki/IGEM:Brown/2007 2007 Brown iGEM team]. The schematic process is shown below.
The Experiments
We tested out detectors (protocol can be found here) and measured the sensitivity using a spectrophotometer. We measured in Flu./OD which is the amount of fluorescence per OD (at 600nm). At first, we encountered many problems. We noticed that all of our dilutions, including the control were approximately the same. We realized that LB (Luria Broth, our growth medium) fluoresces naturally; hence, it masked the effects of the GFP. To solve this problem, we tried to run our tests using M9 Salts as a minimal growth medium. However, our bacteria were unable to grow in the medium. After this, our solution was to grow in LB, spin down the bacteria, then resuspend in PBS (phosphate buffer solution). Lastly, our spectrophotometer still continued to show the same fluorescence reading for most samples except the control. We then realized that the readings had maxed. Serial dilutions were used to determine accurate measurements.
 "
"