Team:Tianjin/Project/Regulation
From 2012.igem.org
Austinamens (Talk | contribs) (→Results and Analysis) |
|||
| (27 intermediate revisions not shown) | |||
| Line 66: | Line 66: | ||
</div> | </div> | ||
<div class="menu_children"> | <div class="menu_children"> | ||
| - | <a href="https://2012.igem.org/Team:Tianjin/Project/Gene"> | + | <a href="https://2012.igem.org/Team:Tianjin/Project/Gene">Future Work</a> |
| + | </div> | ||
| + | <div class="menu_children"> | ||
| + | <a href="https://2012.igem.org/Team:Tianjin/Data">BioBrick</a> | ||
| + | </div> | ||
| + | <div class="menu_children"> | ||
| + | <a href="https://2012.igem.org/Team:Tianjin/Project/Technology">Technology</a> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 82: | Line 88: | ||
<div class="menu_children"> | <div class="menu_children"> | ||
<a href="#Logic_Metabolism_Regulation">Logic Metabolism Regulation</a> | <a href="#Logic_Metabolism_Regulation">Logic Metabolism Regulation</a> | ||
| + | </div> | ||
| + | <div class="menu_children"> | ||
| + | <a href="#References">References</a> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 103: | Line 112: | ||
If we add our O-Key into the expression system, a novel regulation means will exist on the level of translation. Meanwhile, we can combine O-Key with the traditional transcription regulation to create logic gate regulation structure like “AND” gate. The logic gate working with normal close and normal open promoter can constitute multiple “AND/OR/NOR” logic gate control system which has more simple structure compared to traditional regulation. | If we add our O-Key into the expression system, a novel regulation means will exist on the level of translation. Meanwhile, we can combine O-Key with the traditional transcription regulation to create logic gate regulation structure like “AND” gate. The logic gate working with normal close and normal open promoter can constitute multiple “AND/OR/NOR” logic gate control system which has more simple structure compared to traditional regulation. | ||
| - | === | + | ===AND Gate=== |
| - | + | [[file:TJU2012-Proj-LMR-fig-14.png|thumb|300px|center|'''Figure 2.''' Theory figure of logic gate metabolic regulation(from TJU iGEM Team 2012)]] | |
| - | [[file:TJU2012-Proj-LMR-fig- | + | Biological logic gate has been described as the most advanced “biological circuit” ever built. This year, we used the orthogonal system to achieve metabolic regulation by logic gate – the O-Key. Our technology is a translational regulation. It not only introduce a new “AND gate” regulation to cells, but also works perfect with transcriptional regulation, waste no sequence resources and has certain potential of encryption. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The core of our O-Key consists of two part: the o-ribosome and o-mRNA. Their roles in metabolic regulation can be described as key and lock. O-ribosome serves as key, while o-mRNA is the lock. We use the o-RBS to “lock” the target sequence, and make it decipherable only under o-ribosome. They altogether forms an AND gate. | |
| - | + | ||
| - | + | ||
| - | + | Until here, we know that our O-Key system is an AND gate which needs O-Key and O-Lock at the same time to open to express the protein. So we must introduce the O-Key also O-Lock into our system. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[file:TJU2012-Proj-LMR-fig-16.png|thumb|400px|center|'''Figure 3.''' The method to introduce the O-Lock(from TJU iGEM Team 2012)]] | |
| - | + | [[file:TJU2012-Proj-LMR-fig-15.png|thumb|400px|center|'''Figure 4.''' The method to introduce the O-Key(from TJU iGEM Team 2012)]] | |
| - | [[file:TJU2012-Proj-LMR-fig- | + | |
| - | == | + | =<span style="line-height:100%">Synthesizing the Pathway Needed for Synthesizing Violacein</span>= |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===Background=== | ===Background=== | ||
In order to verify the abilities of orthogonal system to adjust metabolism, we chose the metabolic pathway of Violacein. The reason why we chose this one are based on the facts that the pathway is suitable to adjust, and has been deeply learned. | In order to verify the abilities of orthogonal system to adjust metabolism, we chose the metabolic pathway of Violacein. The reason why we chose this one are based on the facts that the pathway is suitable to adjust, and has been deeply learned. | ||
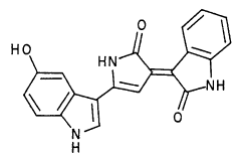
Violacei, the major pigment produced by Chromobacterium violaceum, is a bactericide, a trypanocide, a tumoricide and in addition it has anti-viral activity. | Violacei, the major pigment produced by Chromobacterium violaceum, is a bactericide, a trypanocide, a tumoricide and in addition it has anti-viral activity. | ||
| - | [[file:TJU2012-Proj-LMR-fig-7.png|thumb|500px|center|'''Figure | + | [[file:TJU2012-Proj-LMR-fig-7.png|thumb|500px|center|'''Figure 5.''' Violacein's structure (from "Production, extraction and purification of violacein: an antibiotic pigment produced by Chromobacterium violaceum")]] |
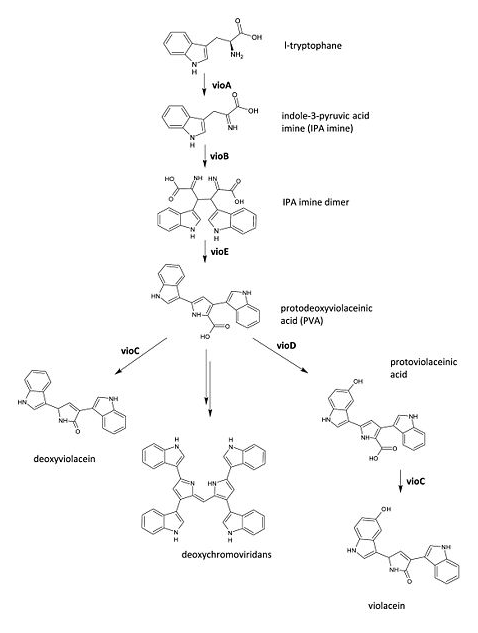
The metabolic pathways to produce Violacein are revealed in the following picture. | The metabolic pathways to produce Violacein are revealed in the following picture. | ||
| - | [[file:TJU2012-Proj-LMR-fig-8.png|thumb| | + | [[file:TJU2012-Proj-LMR-fig-8.png|thumb|300px|center|'''Figure 6.''' Pathway of Violacein (from BBa_K274002)]] |
From the pathway, we can know that except for the desire product, Violacein, the pathway will also produce side product, deoxyviolacein. | From the pathway, we can know that except for the desire product, Violacein, the pathway will also produce side product, deoxyviolacein. | ||
| Line 143: | Line 138: | ||
===Experiment=== | ===Experiment=== | ||
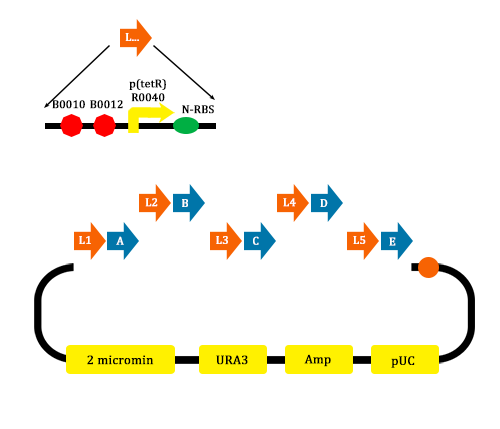
#MAGE mutate the RBS of the gene coding VioD to o-RBS | #MAGE mutate the RBS of the gene coding VioD to o-RBS | ||
| - | #We construct different parts of Vio operons, and transformed into yeast for assembly(In this part, we also assembly the original genes without mutation)[[file:TJU2012-Proj-LMR-fig-9.png|thumb|500px|center|'''Figure | + | #We construct different parts of Vio operons, and transformed into yeast for assembly(In this part, we also assembly the original genes without mutation)[[file:TJU2012-Proj-LMR-fig-9.png|thumb|500px|center|'''Figure 7.''' Assembler of Violacein (from TJU iGEM Team 2012)]] |
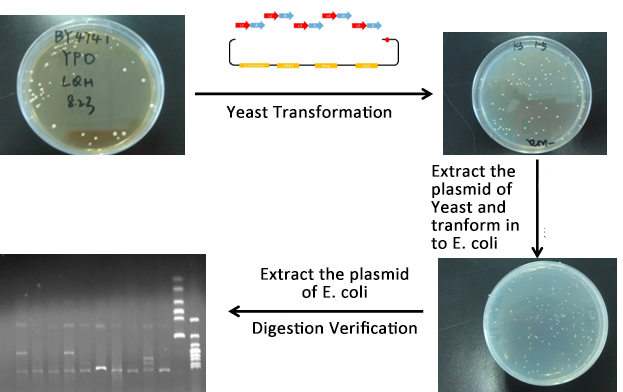
#Extracted the plasmid of Yeast and then transformed into our O-E.coli on LB plates. | #Extracted the plasmid of Yeast and then transformed into our O-E.coli on LB plates. | ||
#Verify through Cpcr and inculcated the right colony into liquid LB overnight. | #Verify through Cpcr and inculcated the right colony into liquid LB overnight. | ||
| Line 149: | Line 144: | ||
===Results and Analysis=== | ===Results and Analysis=== | ||
| - | [[file:TJU2012-Proj-LMR-fig-10.png|thumb|500px|center|'''Figure | + | [[file:TJU2012-Proj-LMR-fig-10.png|thumb|500px|center|'''Figure 8.''' Process of experiments (from TJU iGEM Team 2012)]] |
| - | #From the above picture, the process is finished smoothly and the result of Digested verification is right.[[file:TJU2012-Proj-LMR-fig-11.png|thumb|500px|center|'''Figure | + | #From the above picture, the process is finished smoothly and the result of Digested verification is right.[[file:TJU2012-Proj-LMR-fig-11.png|thumb|500px|center|'''Figure 9.''' Results of the normal pathway (from TJU iGEM Team 2012)]] |
| - | #From this picture, we could find that the test tube with the normal pathway, the liquid is purple. In such condition, for PVA have the larger potential to turn into violacein, violacein stand for a large part and liquid is purple. | + | #From this picture, we could find that the test tube with the normal pathway, the liquid is purple. In such condition, for PVA have the larger potential to turn into violacein, violacein stand for a large part and liquid is purple.[[file:TJU2012-Proj-LMR-fig-12.png|thumb|500px|center|'''Figure 10.''' Results of the mutated pathway without adding Ala (from TJU iGEM Team 2012)]] |
| - | [[file:TJU2012-Proj-LMR-fig-12.png|thumb|500px|center|'''Figure | + | #In this picture, we could find that liquid is blue. As we didn’t add Ala into the liquid, the cells didn’t produce O-ribosome, then the vioD could not be expressed. The PVA could only become deoxyviolacein, which is blue. Therefore the liquid is blue.[[file:TJU2012-Proj-LMR-fig-13.png|thumb|500px|center|'''Figure 11.''' Results of the mutated pathway adding Ala (from TJU iGEM Team 2012)]] |
| - | #In this picture, we could find that liquid is blue. As we didn’t add Ala into the liquid, the cells didn’t produce O-ribosome, then the vioD could not be expressed. The PVA could only become deoxyviolacein, which is blue. Therefore the liquid is blue.[[file:TJU2012-Proj-LMR-fig-13.png|thumb|500px|center|'''Figure | + | |
#In this picture, the liquid is lavender. For this tube, we added Ala, which could induce the production of O-ribosome, then PVA could turn into violacein, then the liquid is lavender. | #In this picture, the liquid is lavender. For this tube, we added Ala, which could induce the production of O-ribosome, then PVA could turn into violacein, then the liquid is lavender. | ||
#From the results of this experiment, we could find that the orthogonal system could work well when used to adjust metabolism, and the effect is satisfied. In the fourth part, we will further discuss the application of the orthogonal system in regulating the metabolism. | #From the results of this experiment, we could find that the orthogonal system could work well when used to adjust metabolism, and the effect is satisfied. In the fourth part, we will further discuss the application of the orthogonal system in regulating the metabolism. | ||
=Logic Gate Metabolic Regulation= | =Logic Gate Metabolic Regulation= | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
We use modeling fitting to predict the key nodes in the metabolism network. MAGE can be used to mutate the RBS of target gene to o-RBS thus locking the target gene. Compared with the conventional way of regulating by overexpression and gene knockout, our O-Key avoids decreasing cell activity and slowing growth rate because of partial adjustment of regulation networks by altering multiple cell native control transcription simultaneously. This technology saves time and boost efficiency. | We use modeling fitting to predict the key nodes in the metabolism network. MAGE can be used to mutate the RBS of target gene to o-RBS thus locking the target gene. Compared with the conventional way of regulating by overexpression and gene knockout, our O-Key avoids decreasing cell activity and slowing growth rate because of partial adjustment of regulation networks by altering multiple cell native control transcription simultaneously. This technology saves time and boost efficiency. | ||
| - | + | ||
| + | <span style="font-size:12px"><center>'''Table 1.''' The difference between original regulation and orthogonal regulation (from TJU iGEM Team 2012)</center></span> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <table border="0" | ||
| + | cellpadding="5" | ||
| + | style="border:2px solid #7b3101;"> | ||
| + | |||
| + | <tr> | ||
| + | <th style="background:#fbb181;">O-Key (Orthogonal regulation)</th> | ||
| + | <th style="background:#fbb181;">Conventional Regulation (Overexpression, gene knockout)</th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td style="background:#fdd8c0;">Integrted regulation; Easy to operate and combine with model fitting analysis</td> | ||
| + | <td style="background:#fdd8c0;">Partial adjustment<br>Laborious, time-consuming, complicated</td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | </center> | ||
| + | </html> | ||
We will elaborate on the function of O-Key in the following examples. | We will elaborate on the function of O-Key in the following examples. | ||
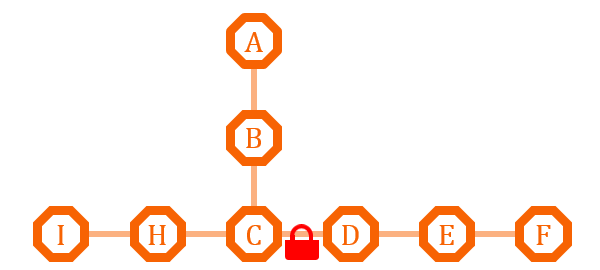
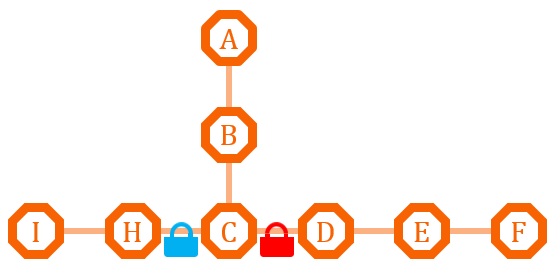
| - | [[ | + | [[File:TJU2012-Proj-LMR-fig-18.png|thumb|500px|center|'''Figure 12.''' The first example of metabolism regulation (from TJU iGEM Team 2012)]] |
This pathway has two branches. If we put an O-Lock between C and D, we can adjust the metabolic pathway according to the cell growth cycle. Say I is the primary metabolites, and the bacteria need it for growth, we can close the O-Lock to shut down pathway D. When the bacteria is in stationary phase and in sufficient quantity, we can open the O-Lock to turn on pathway D, and increase the secondary metabolites F by its competitive advantage. | This pathway has two branches. If we put an O-Lock between C and D, we can adjust the metabolic pathway according to the cell growth cycle. Say I is the primary metabolites, and the bacteria need it for growth, we can close the O-Lock to shut down pathway D. When the bacteria is in stationary phase and in sufficient quantity, we can open the O-Lock to turn on pathway D, and increase the secondary metabolites F by its competitive advantage. | ||
| - | [[file:TJU2012-Proj-LMR-fig-19.png|thumb|500px|center|'''Figure | + | [[file:TJU2012-Proj-LMR-fig-19.png|thumb|500px|center|'''Figure 13.''' The second example of metabolism regulation (from TJU iGEM Team 2012)]] |
If the natural branch competitive advantage does not exist, or we need precise regulation of the metabolism of the two pathway, we can install different O-Key system on the two pathway, and use the O-Key to achieve accurate control. | If the natural branch competitive advantage does not exist, or we need precise regulation of the metabolism of the two pathway, we can install different O-Key system on the two pathway, and use the O-Key to achieve accurate control. | ||
In a word, the O-Key system has two advantages. On the one hand, it can achieve the same function of other complex operons using much more concise sequence. | In a word, the O-Key system has two advantages. On the one hand, it can achieve the same function of other complex operons using much more concise sequence. | ||
| - | [[file:TJU2012-Proj-LMR-fig-20.png|thumb|500px|center|'''Figure | + | [[file:TJU2012-Proj-LMR-fig-20.png|thumb|500px|center|'''Figure 14.''' One example of logic metabolism regulation (from "Synthesis of orthogonal transcription-translation networks")]] |
Moreover, with the combination of different O-Key system we are able to construct more complicated logic networks using in a much simpler way. Currently, we have demonstrated that bacteria and DNA molecules can “copy” logic gate, and started constructing more complicated logic gates. | Moreover, with the combination of different O-Key system we are able to construct more complicated logic networks using in a much simpler way. Currently, we have demonstrated that bacteria and DNA molecules can “copy” logic gate, and started constructing more complicated logic gates. | ||
| - | We wish this research would result in a new generation biological processor, and they have the same importance in information processing as other electric devices. Although there is still a | + | We wish this research would result in a new generation biological processor, and they have the same importance in information processing as other electric devices. Although there is still a |
=References= | =References= | ||
| - | # Daniel G. Gibsona, Gwynedd A. Bendersb, | + | # Lon M. Chubiz and Christopher V. Rao(2008). Computational design of orthogonal ribosomes, vol.36 no.12 |
| - | # Daniel G. Gibson | + | # Daniel G. Gibsona,1, Gwynedd A. Bendersb, Kevin C. Axelrod PNAS(2008). One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome, vol. 105 no. 51 |
| - | # Zengyi Shao, Hua Zhao1and Huimin Zhao | + | # Daniel G. Gibson(2009).Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. vol.37 no.20Zengyi Shao, Hua Zhao1and Huimin Zhao(2009). DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. vol.37 no.2 |
| - | # | + | # Zengyi Shao, Hua Zhao1and Huimin Zhao(2009). DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. vol.37 no.2Daniel G. Gibsona, Gwynedd A. Bendersb, et al., Next-generation synthetic gene network |
| - | + | # Howard M Salis, Ethan A Mirsky & Christopher A Voigt(2009).Automated design of synthetic ribosome binding sites to control protein expression, doi:10.1038/nbt.1568 | |
| - | # Howard M Salis, Ethan A Mirsky & Christopher A Voigt | + | # Howard M Salis, Ethan A Mirsky & Christopher A Voigt(2009).Automated design of synthetic ribosome binding sites to control protein expression, doi:10.1038/nbt.1568 |
| - | # Wenlin An and Jason W. Chinl | + | # Wenlin An and Jason W. Chinl(2009).Synthesis of orthogonal transcriptiontranslation networks vol.106 no.21 |
| - | # | + | # George H.McArthur IV and Stephen S. Fong, Toward Engineering Synthetic Microbial Metabolism |
| - | + | ||
{{:Team:Tianjin/footer}} | {{:Team:Tianjin/footer}} | ||
Latest revision as of 18:57, 26 October 2012

Background
Metabolic Regulation
Cell metabolism is the foundation of cell growth, secretion, differentiation, etc. as well as the core process of the modern fermentation technology that can make large impact on the quality and output of product.
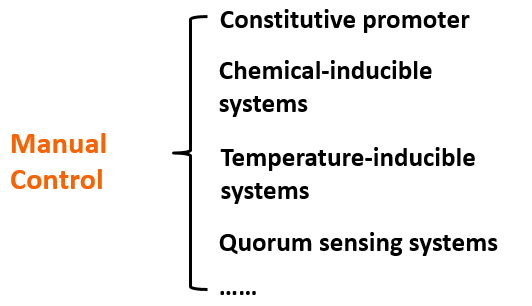
However the modern metabolic regulation of strains has a large amount of areas for improvement. Take the most common E. coli as example. At present, most of the regulation means is to use a single promoter to construct specific operon gene cluster to achieve the regulation with the addition of certain inducers induction of a specific protein. But this induced expression is typically unidirectional, irreversible and it needs to build many complex operons’ gene structure to construct multiple logical regulation, this will produce a plenty of limitation.
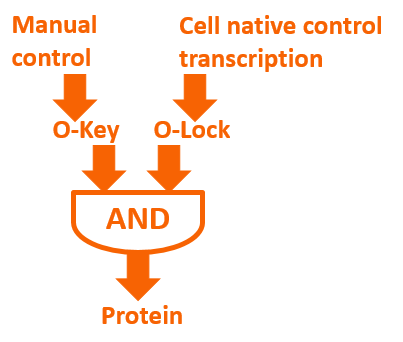
If we add our O-Key into the expression system, a novel regulation means will exist on the level of translation. Meanwhile, we can combine O-Key with the traditional transcription regulation to create logic gate regulation structure like “AND” gate. The logic gate working with normal close and normal open promoter can constitute multiple “AND/OR/NOR” logic gate control system which has more simple structure compared to traditional regulation.
AND Gate
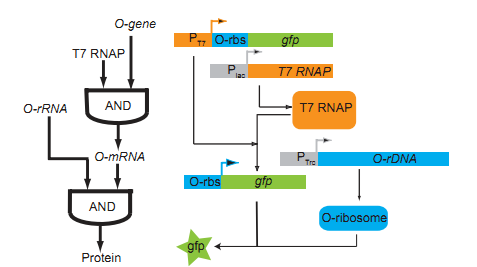
Biological logic gate has been described as the most advanced “biological circuit” ever built. This year, we used the orthogonal system to achieve metabolic regulation by logic gate – the O-Key. Our technology is a translational regulation. It not only introduce a new “AND gate” regulation to cells, but also works perfect with transcriptional regulation, waste no sequence resources and has certain potential of encryption.
The core of our O-Key consists of two part: the o-ribosome and o-mRNA. Their roles in metabolic regulation can be described as key and lock. O-ribosome serves as key, while o-mRNA is the lock. We use the o-RBS to “lock” the target sequence, and make it decipherable only under o-ribosome. They altogether forms an AND gate.
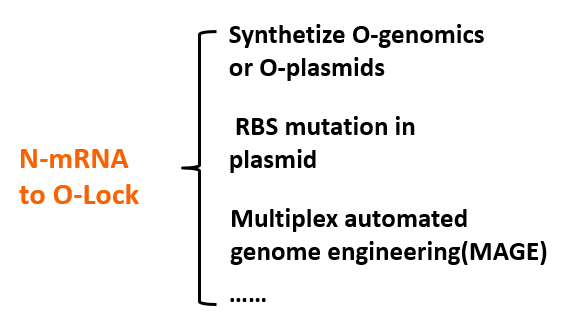
Until here, we know that our O-Key system is an AND gate which needs O-Key and O-Lock at the same time to open to express the protein. So we must introduce the O-Key also O-Lock into our system.
Synthesizing the Pathway Needed for Synthesizing Violacein
Background
In order to verify the abilities of orthogonal system to adjust metabolism, we chose the metabolic pathway of Violacein. The reason why we chose this one are based on the facts that the pathway is suitable to adjust, and has been deeply learned.
Violacei, the major pigment produced by Chromobacterium violaceum, is a bactericide, a trypanocide, a tumoricide and in addition it has anti-viral activity.
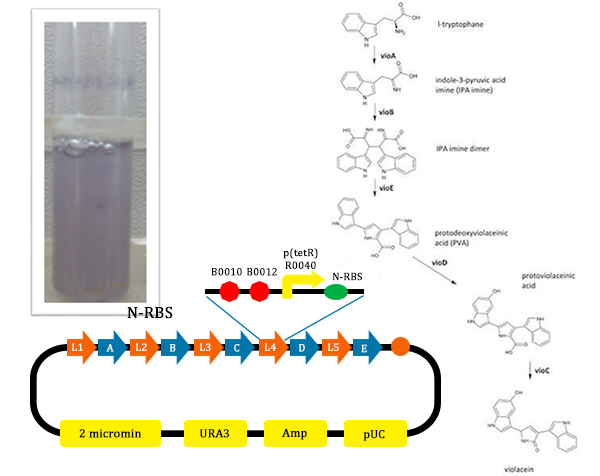
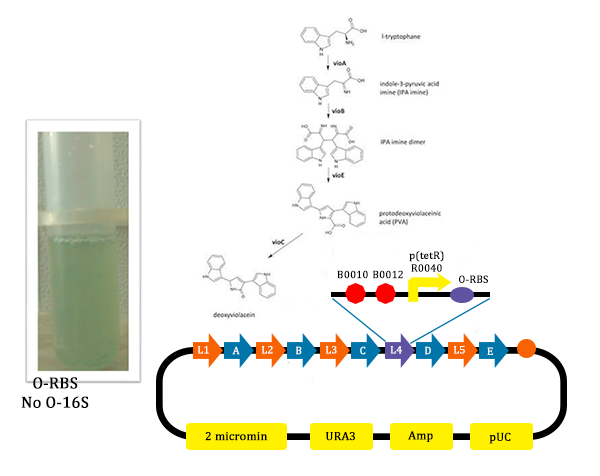
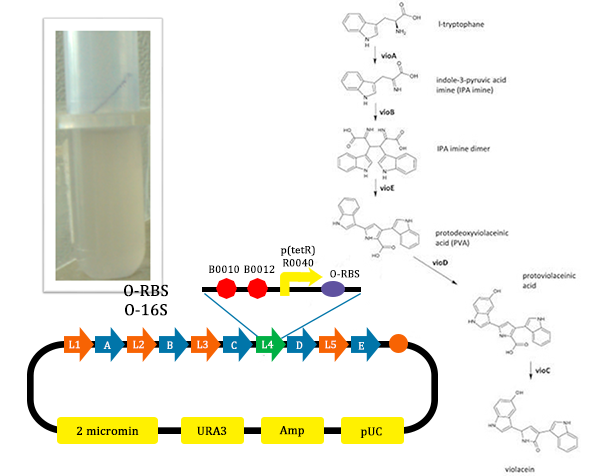
The metabolic pathways to produce Violacein are revealed in the following picture.
From the pathway, we can know that except for the desire product, Violacein, the pathway will also produce side product, deoxyviolacein.
Principle
In order to verify the feasibility of the orthogonal system in adjusting metabolism, we mutated the RBS of the gene encoding VioD by MAGE, and assembled in Yeast Assembler. The VioD gene and the O-Key gene with pBad promoter were finally transformed into the cell. Because the cell itself doesn’t have the O-Key, the gene is stringently shut down. When we added Ala to induce pBad, the O-Key were produced to open the O-Lock, so the cell could produce violacin.
Experiment
- MAGE mutate the RBS of the gene coding VioD to o-RBS
- We construct different parts of Vio operons, and transformed into yeast for assembly(In this part, we also assembly the original genes without mutation)
- Extracted the plasmid of Yeast and then transformed into our O-E.coli on LB plates.
- Verify through Cpcr and inculcated the right colony into liquid LB overnight.
- Extracted the plasmid of E.coli and verified through digestion of ligase.
Results and Analysis
- From the above picture, the process is finished smoothly and the result of Digested verification is right.
- From this picture, we could find that the test tube with the normal pathway, the liquid is purple. In such condition, for PVA have the larger potential to turn into violacein, violacein stand for a large part and liquid is purple.
- In this picture, we could find that liquid is blue. As we didn’t add Ala into the liquid, the cells didn’t produce O-ribosome, then the vioD could not be expressed. The PVA could only become deoxyviolacein, which is blue. Therefore the liquid is blue.
- In this picture, the liquid is lavender. For this tube, we added Ala, which could induce the production of O-ribosome, then PVA could turn into violacein, then the liquid is lavender.
- From the results of this experiment, we could find that the orthogonal system could work well when used to adjust metabolism, and the effect is satisfied. In the fourth part, we will further discuss the application of the orthogonal system in regulating the metabolism.
Logic Gate Metabolic Regulation
We use modeling fitting to predict the key nodes in the metabolism network. MAGE can be used to mutate the RBS of target gene to o-RBS thus locking the target gene. Compared with the conventional way of regulating by overexpression and gene knockout, our O-Key avoids decreasing cell activity and slowing growth rate because of partial adjustment of regulation networks by altering multiple cell native control transcription simultaneously. This technology saves time and boost efficiency.
| O-Key (Orthogonal regulation) | Conventional Regulation (Overexpression, gene knockout) |
|---|---|
| Integrted regulation; Easy to operate and combine with model fitting analysis | Partial adjustment Laborious, time-consuming, complicated |
This pathway has two branches. If we put an O-Lock between C and D, we can adjust the metabolic pathway according to the cell growth cycle. Say I is the primary metabolites, and the bacteria need it for growth, we can close the O-Lock to shut down pathway D. When the bacteria is in stationary phase and in sufficient quantity, we can open the O-Lock to turn on pathway D, and increase the secondary metabolites F by its competitive advantage.
If the natural branch competitive advantage does not exist, or we need precise regulation of the metabolism of the two pathway, we can install different O-Key system on the two pathway, and use the O-Key to achieve accurate control.
In a word, the O-Key system has two advantages. On the one hand, it can achieve the same function of other complex operons using much more concise sequence.
Moreover, with the combination of different O-Key system we are able to construct more complicated logic networks using in a much simpler way. Currently, we have demonstrated that bacteria and DNA molecules can “copy” logic gate, and started constructing more complicated logic gates.
We wish this research would result in a new generation biological processor, and they have the same importance in information processing as other electric devices. Although there is still a
References
- Lon M. Chubiz and Christopher V. Rao(2008). Computational design of orthogonal ribosomes, vol.36 no.12
- Daniel G. Gibsona,1, Gwynedd A. Bendersb, Kevin C. Axelrod PNAS(2008). One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome, vol. 105 no. 51
- Daniel G. Gibson(2009).Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. vol.37 no.20Zengyi Shao, Hua Zhao1and Huimin Zhao(2009). DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. vol.37 no.2
- Zengyi Shao, Hua Zhao1and Huimin Zhao(2009). DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. vol.37 no.2Daniel G. Gibsona, Gwynedd A. Bendersb, et al., Next-generation synthetic gene network
- Howard M Salis, Ethan A Mirsky & Christopher A Voigt(2009).Automated design of synthetic ribosome binding sites to control protein expression, doi:10.1038/nbt.1568
- Howard M Salis, Ethan A Mirsky & Christopher A Voigt(2009).Automated design of synthetic ribosome binding sites to control protein expression, doi:10.1038/nbt.1568
- Wenlin An and Jason W. Chinl(2009).Synthesis of orthogonal transcriptiontranslation networks vol.106 no.21
- George H.McArthur IV and Stephen S. Fong, Toward Engineering Synthetic Microbial Metabolism
 "
"