Team:Colombia/Project/Experiments/Aliivibrio and Streptomyces
From 2012.igem.org
(→Aim) |
(→Aim) |
||
| (2 intermediate revisions not shown) | |||
| Line 56: | Line 56: | ||
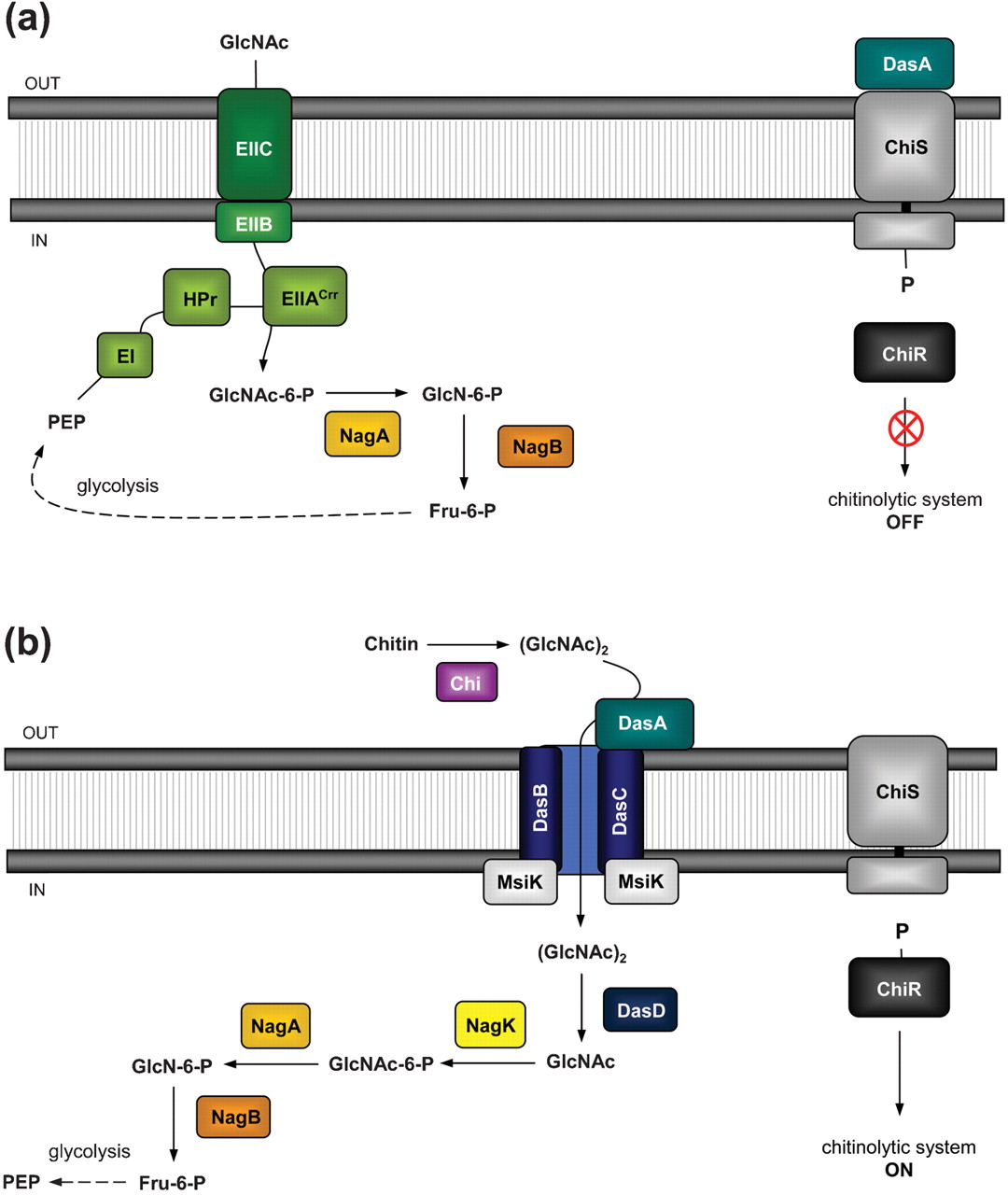
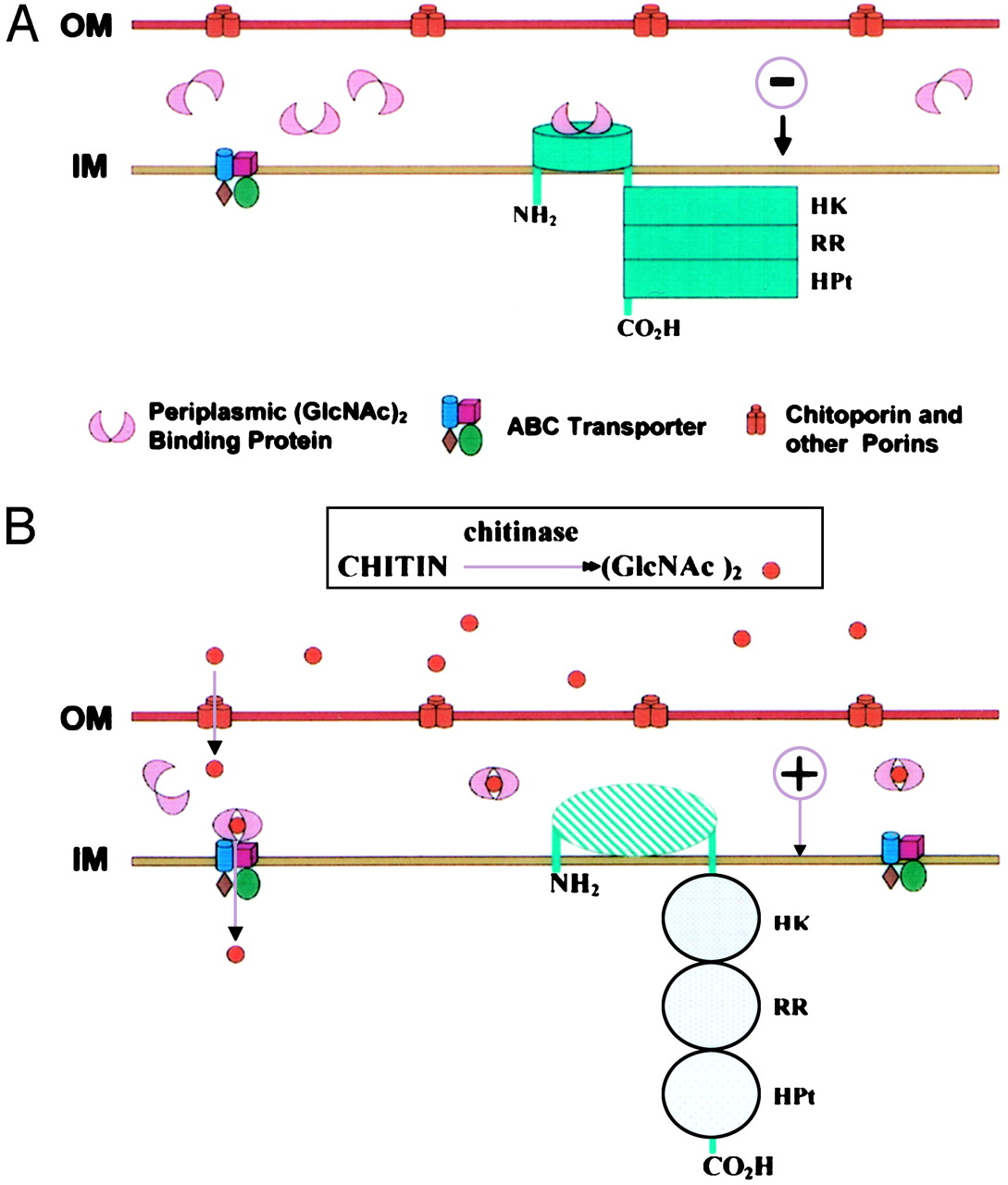
''Vibrio fischeri'' ES114 ([http://ijs.sgmjournals.org/content/57/12/2823 now ''Aliivibrio fischeri'' ES114]) and ''Streptomyces coelicolor'' A3(2) are environmental bacteria with well-characterized detection and catabolic cascades for chitin use as a carbon source. The way by which each bacteria detect the chitin is through a two-component system. '''Figure 1''' illustrates the proposed sensing model for the system in ''A. fischeri''. | ''Vibrio fischeri'' ES114 ([http://ijs.sgmjournals.org/content/57/12/2823 now ''Aliivibrio fischeri'' ES114]) and ''Streptomyces coelicolor'' A3(2) are environmental bacteria with well-characterized detection and catabolic cascades for chitin use as a carbon source. The way by which each bacteria detect the chitin is through a two-component system. '''Figure 1''' illustrates the proposed sensing model for the system in ''A. fischeri''. | ||
| - | Chitin is fragmented into N-acetyl glucosamine (GlcNAc)2 dimers through the action of a chitinase ('''ChiA'''). (GlcNAc)2 passes to the periplasmic space through chitoporin ('''ChiP''') and binds the chitin-binding protein ('''CBP''') which leads to the signalling from '''ChiS''' histidine kinase to turn on the chitinolytic genes. '''Figure 2''' depicts how ''S. coelicolor'' uses a pretty similar system to the previously described | + | Chitin is fragmented into N-acetyl glucosamine (GlcNAc)2 dimers through the action of a chitinase ('''ChiA'''). (GlcNAc)2 passes to the periplasmic space through chitoporin ('''ChiP''') and binds the chitin-binding protein ('''CBP''') which leads to the signalling from '''ChiS''' histidine kinase to turn on the chitinolytic genes. |
| + | |||
| + | |||
| + | '''Figure 2''' depicts how ''S. coelicolor'' uses a pretty similar system to the previously described. The analog channel that allows the (GlcNAc)2 to cross the plasmatic membrane. (GlcNAc)2-activated '''DasA''' serves as an activator to '''ChiS''', which phosphorylates '''ChiR''' regulator to induce transcription of the chitin metabolism genes. | ||
In both cases, we're working on to extract and put the genes into functional and independent biobricks available to further applications of chitin sensing and/or degradation. | In both cases, we're working on to extract and put the genes into functional and independent biobricks available to further applications of chitin sensing and/or degradation. | ||
Latest revision as of 03:04, 27 September 2012
Template:Https://2012.igem.org/User:Tabima
Contents |
Aim
Our system heavily relies in the ability of our bacteria to sense environmental cues belonging to the pathogens we’re particularly concerned of.
Chitin is one of the main components of fungal cell walls and, in consequence, we will use it as an indicator of fungal infection.
Vibrio fischeri ES114 ([http://ijs.sgmjournals.org/content/57/12/2823 now Aliivibrio fischeri ES114]) and Streptomyces coelicolor A3(2) are environmental bacteria with well-characterized detection and catabolic cascades for chitin use as a carbon source. The way by which each bacteria detect the chitin is through a two-component system. Figure 1 illustrates the proposed sensing model for the system in A. fischeri.
Chitin is fragmented into N-acetyl glucosamine (GlcNAc)2 dimers through the action of a chitinase (ChiA). (GlcNAc)2 passes to the periplasmic space through chitoporin (ChiP) and binds the chitin-binding protein (CBP) which leads to the signalling from ChiS histidine kinase to turn on the chitinolytic genes.
Figure 2 depicts how S. coelicolor uses a pretty similar system to the previously described. The analog channel that allows the (GlcNAc)2 to cross the plasmatic membrane. (GlcNAc)2-activated DasA serves as an activator to ChiS, which phosphorylates ChiR regulator to induce transcription of the chitin metabolism genes.
In both cases, we're working on to extract and put the genes into functional and independent biobricks available to further applications of chitin sensing and/or degradation.
Our bacteria
Aliivibrio fischeri ES114
| Aliivibrio fischeri ES114 | ||
|---|---|---|
| Taxonomy[1] | Superkingdom | Bacteria |
| Phylum | Proteobacteria | |
| Class | Gammaproteobacteria | |
| Order | Vibrionales | |
| Family | Vibrionaceae | |
| Genus | Aliivibrio | |
| Species | A. fischeri | |
| Strain | ES114 | |
We're extracting the whole chitin sensing system, namely [http://www.ncbi.nlm.nih.gov/gene/3279287 chiS], [http://www.ncbi.nlm.nih.gov/gene/3279194 chiP], [http://www.ncbi.nlm.nih.gov/gene/3277358 chiA] and [http://www.ncbi.nlm.nih.gov/gene/3279341 CBP]. Also, as the promoter that responses to the signalling from chiS is not known, we're also extracting the 100kb upstream region of chiP that we named pChitin.
Streptomyces coelicolor A3(2)
| Streptomyces coelicolor A3(2) | ||
|---|---|---|
| Taxonomy[2] | Superkingdom | Bacteria |
| Phylum | Actinobacteria | |
| Class | Actinobacteria | |
| Order | Actinomycetales | |
| Family | Streptomycetaceae | |
| Genus | Streptomyces | |
| Species | Streptomyces coelicolor | |
| Strain | A3(2) | |
From this bacterium, we're planning the cloning of the operons [http://www.ncbi.nlm.nih.gov/gene?term=SCO5377 chiSR], [http://www.ncbi.nlm.nih.gov/gene/1100673 dasABC], the genes [http://www.ncbi.nlm.nih.gov/gene/1099680 msiK], [http://www.ncbi.nlm.nih.gov/gene?term=SCO5376 chiC] and the promoter pChitSc, which corresponds to the 60bp before chiC, previously reported in [5].
References
[1] [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=312309&lvl=3&lin=f&keep=1&srchmode=1&unlock NCBI Taxonomy Browser: Aliivibrio fischeri ES114]
[2] [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=312309&lvl=3&lin=f&keep=1&srchmode=1&unlock NCBI Taxonomy Browser: Streptomyces coelicolor A3(2)]
[3] [http://www.pnas.org/content/101/2/627 Li, X., and Roseman, S. (2004). The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. PNAS 101, 627–631.]
[4] [http://mic.sgmjournals.org/content/154/2/373.full Colson, S., Wezel, G.P. van, Craig, M., Noens, E.E.E., Nothaft, H., Mommaas, A.M., Titgemeyer, F., Joris, B., and Rigali, S. (2008). The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154, 373–382.]
[5] [http://www.springerlink.com/content/fh6g740578v78277/ Homerová, D., Knirschová, R., and Kormanec, J. (2002). Response regulator ChiR regulates expression of chitinase gene, chiC, in Streptomyces coelicolor. Folia Microbiol. (Praha) 47, 499–505.]
 "
"