Team:Potsdam Bioware/Project/Part Antibody
From 2012.igem.org
(→Immunfluorescence) |
(→Exchange of Signal Peptide and Transmembrane Domain) |
||
| (64 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| - | <div class="box_round | + | <div class="box_round white_bg"> |
| - | + | ||
| - | + | ||
==Antibody Module== | ==Antibody Module== | ||
| Line 18: | Line 16: | ||
==== A Short Review on Antibodies ==== | ==== A Short Review on Antibodies ==== | ||
Antibodies are highly specific targeting reagents and are the key defense system for recognizing pathogens and toxins. Antibodies are Y shaped multi domain proteins with two antigen binding sites displaying the Fab fragment and one effector domain represented by the Fc domain. (Holliger, Hudson; 2005) The Fab fragment is composed of two antigen recognition sites with one variable light (VL) and constant light (VC) chain and one variable heavy (VH) and constant heavy (CH) chain each which allow the specific and affine binding of antigens. By interacting with the complement and the Fc receptor, the Fc domain is able to mediate cytotoxic effector functions. (Holliger, Hudson; 2005) | Antibodies are highly specific targeting reagents and are the key defense system for recognizing pathogens and toxins. Antibodies are Y shaped multi domain proteins with two antigen binding sites displaying the Fab fragment and one effector domain represented by the Fc domain. (Holliger, Hudson; 2005) The Fab fragment is composed of two antigen recognition sites with one variable light (VL) and constant light (VC) chain and one variable heavy (VH) and constant heavy (CH) chain each which allow the specific and affine binding of antigens. By interacting with the complement and the Fc receptor, the Fc domain is able to mediate cytotoxic effector functions. (Holliger, Hudson; 2005) | ||
| - | ===== Single Chain Fragment Variable ===== | + | ===== Single Chain Fragment Variable Domain===== |
A single chain fragment variable is a small unit of immunoglobulin and consists of the variable heavy (VH) and the variable light (VL) domains which are joined together by a flexible peptide linker. (Alitheen and Hamid et al.; 2012) The scFv is a very popular format that shows a high antigen binding capacity and can be easily expressed. | A single chain fragment variable is a small unit of immunoglobulin and consists of the variable heavy (VH) and the variable light (VL) domains which are joined together by a flexible peptide linker. (Alitheen and Hamid et al.; 2012) The scFv is a very popular format that shows a high antigen binding capacity and can be easily expressed. | ||
<br> | <br> | ||
| - | [[file: | + | [[file:UP12_Antibody_overview.png|center|400px|thumb|'''Figure 1:''' Selected antibody forms relative to therapeutic applications]] |
<br> | <br> | ||
| Line 33: | Line 31: | ||
The scFv425 is of murine origin and was derived against the human epidermal growth factor receptor domain 3. It was originally described in context of a publication dealing with immunotherapy of pancreatic carcinoma cells which consequently express the EGFR. The scFv was used as mediator of carcinoma cells and an endotoxin, thus showing the high relevance of the single chain fragment for medical purposes. (Bruell et al; 2005) | The scFv425 is of murine origin and was derived against the human epidermal growth factor receptor domain 3. It was originally described in context of a publication dealing with immunotherapy of pancreatic carcinoma cells which consequently express the EGFR. The scFv was used as mediator of carcinoma cells and an endotoxin, thus showing the high relevance of the single chain fragment for medical purposes. (Bruell et al; 2005) | ||
<br> | <br> | ||
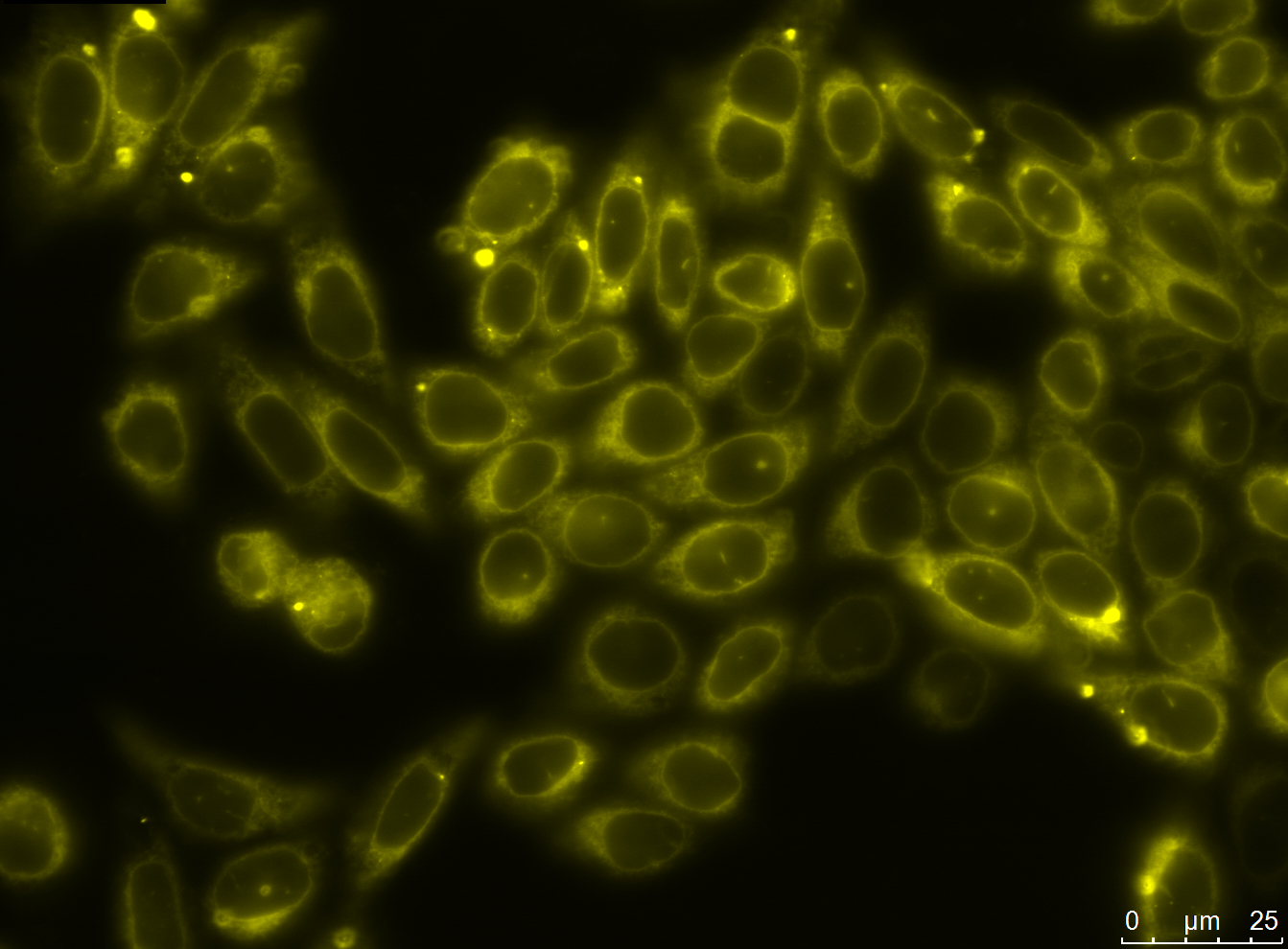
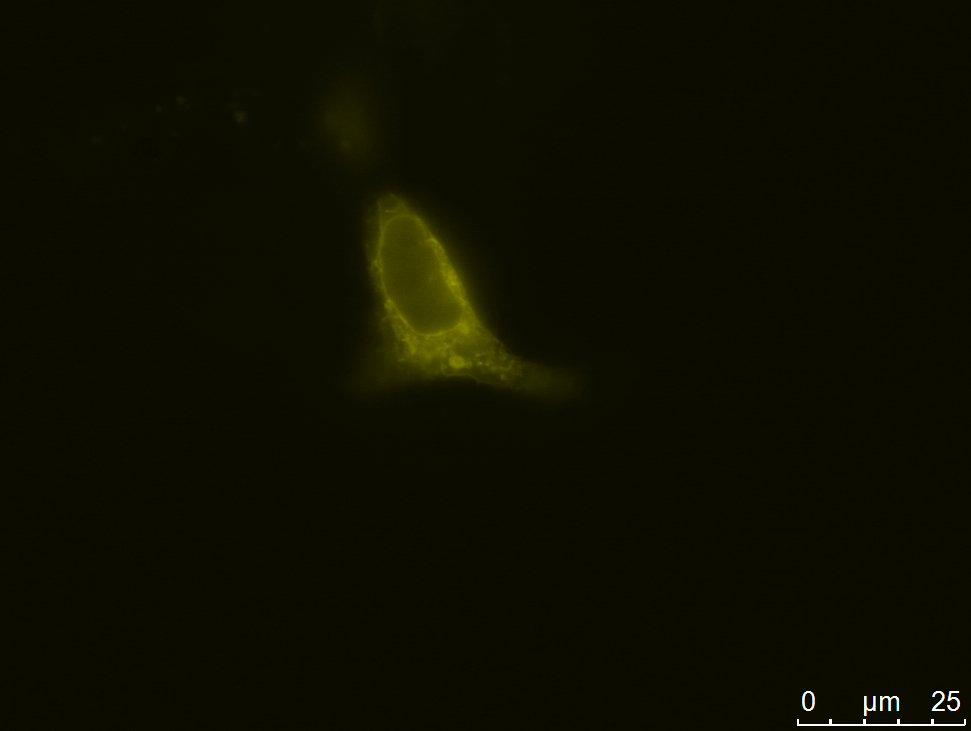
| - | [[file:UP12_smaller-antibody_construct.jpg|center|300px|thumb| | + | [[file:UP12_smaller-antibody_construct.jpg|center|300px|thumb|'''Figure 2:''' Localization of the smaller antibody construct in transient transfected CHO cells]] |
| - | + | ||
===== Advanced Antibody Construct ===== | ===== Advanced Antibody Construct ===== | ||
| Line 41: | Line 38: | ||
Additionallly we integrated a switch on the protein level which is represented by the TEV protease cleavage site permitting the shift to a secretory antibody production. Expression of the antibody construct and its cellular localization can be monitored by the mCherry signal. | Additionallly we integrated a switch on the protein level which is represented by the TEV protease cleavage site permitting the shift to a secretory antibody production. Expression of the antibody construct and its cellular localization can be monitored by the mCherry signal. | ||
<br> | <br> | ||
| - | [[file:UP12_advanced_antibody_construct.jpg|center|300px|thumb| | + | [[file:UP12_advanced_antibody_construct.jpg|center|300px|thumb|'''Figure 3:''' Localization of the advanced antibody construct in transient transfected CHO cells]] |
<br> | <br> | ||
| Line 47: | Line 44: | ||
Chinese hamster ovary cells (CHO) are an immortal mammalian cell line which is frequently used for industrial recombinant protein expressions. Problems when working with mammalian cells are the varying transfection rate and the transitory nature of transient transfected plasmids. To overcome this problem we are working with a CHO expression cell line for the stable transfection of our construct of interest by using Flp recombinase-mediated integration. | Chinese hamster ovary cells (CHO) are an immortal mammalian cell line which is frequently used for industrial recombinant protein expressions. Problems when working with mammalian cells are the varying transfection rate and the transitory nature of transient transfected plasmids. To overcome this problem we are working with a CHO expression cell line for the stable transfection of our construct of interest by using Flp recombinase-mediated integration. | ||
<br> | <br> | ||
| - | [[file:UP12_CHO_cells.jpg|center|300px|thumb| | + | [[file:UP12_CHO_cells.jpg|center|300px|thumb|'''Figure 4:''' CHO cells in culture]] |
<br> | <br> | ||
| Line 53: | Line 50: | ||
The FRT/Flp system (LifeTechnologies) is based on homologous recombination initiated by the enzyme flippase (Flp) at a specific FRT site in the genome. CHO cell containing a randomly but stably integrated FRT sequence can be selected by antibiotic resistance against Zeocin. Another FRT sequence is located on the shuttle plasmid (pcDNA/5FRT) that contains the gene of interest and a second antibiotic resistance against Hygromycin. The system is accomplished by the third plasmid (pOG44) carrying the recombinase flippase (Flp) which mediates the recombination of both FRT sequences leading to an exchange of Zeocin to Hygromycin resistance. | The FRT/Flp system (LifeTechnologies) is based on homologous recombination initiated by the enzyme flippase (Flp) at a specific FRT site in the genome. CHO cell containing a randomly but stably integrated FRT sequence can be selected by antibiotic resistance against Zeocin. Another FRT sequence is located on the shuttle plasmid (pcDNA/5FRT) that contains the gene of interest and a second antibiotic resistance against Hygromycin. The system is accomplished by the third plasmid (pOG44) carrying the recombinase flippase (Flp) which mediates the recombination of both FRT sequences leading to an exchange of Zeocin to Hygromycin resistance. | ||
<br> | <br> | ||
| - | [[file:UP12_Flp-In-system.jpg|center|900px|thumb| | + | [[file:UP12_Flp-In-system.jpg|center|900px|thumb|'''Figure 5:''' Flp-In system of LifeTechnologies applied with our sequences in CHO cells]] |
<br> | <br> | ||
Both of our antibody constructs were also transfected transiently to get a first impression of the expression level, the cellular localization and the compatibility with the CHO cell metabolism. | Both of our antibody constructs were also transfected transiently to get a first impression of the expression level, the cellular localization and the compatibility with the CHO cell metabolism. | ||
| Line 67: | Line 64: | ||
===== Sensitivity to Hygromycin for Flp-In System ===== | ===== Sensitivity to Hygromycin for Flp-In System ===== | ||
| - | After 48 hours of co-transfection of CHO cells with pOG 44 and the Flp-in expression construct we selected stable transfected cells by using | + | After 48 hours of co-transfection of CHO cells with pOG 44 and the Flp-in expression construct we selected stable transfected cells by using Hygromycin B. |
| - | Before we started with transfection we determined the optimal | + | Before we started with transfection we determined the optimal Hygromycin concentration by testing the sensitivity of the CHO Flp-In cells. |
| - | For this purpose we applied different | + | For this purpose we applied different Hygromycin concentrations over a period of 6 days and determined cell numbers by counting cells every 24h. |
| - | Since | + | Since Hygromycin only affects growing cells we looked for the concentration at which cell growth was prevented. According to the data shown in Figure 6 we decided to carry out the selection with a Hygromycin concentration of 550 µg/ml. |
| - | [[file:UP12_hygromycin_sensitivity.jpg|center| | + | [[file:UP12_hygromycin_sensitivity.jpg|center|700px|thumb|'''Figure 6:''' Determination of optimal Hygromycin concentration for stable transfection of CHO Flp-In cells.]] |
<br> | <br> | ||
| Line 79: | Line 76: | ||
The sequence from the human EGFR (ErbB-1) signal peptide was taken from the already existing BioBrick BBa_K157001 and was fused N-terminally to the scFv 425 against the epidermal growth factor receptor (EGFR) domain 3. The scFv425 has a shortened N-terminal FLAG-tag sequence of five amino acids (DYKDE) that allows us a handy purification and simplifies its detection. A TEV protease cleavage site was integrated between the scFv and the transmembrane domain by primer extension and permits the shift from surface presentation to secretion on protein level. The TEV protease recognition site ENLYFQG was used here and represents the most commonly used aa sequence for recognition by the 27kDA catalytic domain of Nuclear Inclusion a (NIa) protein encoded by the tobacco etch virus (TEV). For an unimpeded activity of the TEV protease a three amino acid linker at the N-terminal end and a two amino acid linker at the C-terminal end were added to the sequence. The helical single-span transmembrane domain of the B-cell receptor (BBa_K157010) was modified in terms of the shortening of the linker sequence to three amino acids at the C-terminal end. Expression of the scFv fusion protein and its cellular localization can be monitored by the enhanced YFP (BBa_E0030) signal with an excitation at 514 nm and an emission at 527 nm. | The sequence from the human EGFR (ErbB-1) signal peptide was taken from the already existing BioBrick BBa_K157001 and was fused N-terminally to the scFv 425 against the epidermal growth factor receptor (EGFR) domain 3. The scFv425 has a shortened N-terminal FLAG-tag sequence of five amino acids (DYKDE) that allows us a handy purification and simplifies its detection. A TEV protease cleavage site was integrated between the scFv and the transmembrane domain by primer extension and permits the shift from surface presentation to secretion on protein level. The TEV protease recognition site ENLYFQG was used here and represents the most commonly used aa sequence for recognition by the 27kDA catalytic domain of Nuclear Inclusion a (NIa) protein encoded by the tobacco etch virus (TEV). For an unimpeded activity of the TEV protease a three amino acid linker at the N-terminal end and a two amino acid linker at the C-terminal end were added to the sequence. The helical single-span transmembrane domain of the B-cell receptor (BBa_K157010) was modified in terms of the shortening of the linker sequence to three amino acids at the C-terminal end. Expression of the scFv fusion protein and its cellular localization can be monitored by the enhanced YFP (BBa_E0030) signal with an excitation at 514 nm and an emission at 527 nm. | ||
<br> | <br> | ||
| - | [[file:UP12_BBa_K929101.png| | + | [[file:UP12_BBa_K929101.png|left|650px|thumb|'''Figure 7:''' Smaller antibody construct cloned into pSB1C3 (BBa_K929101)]] |
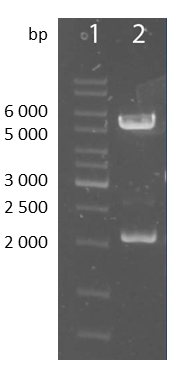
| - | + | [[file:UP12_scFv-cloningpart.png|center|150px|thumb|'''Figure 8:''' Test digestion of the smaller construct with NheI and ApaI; 1: marker; 2: smaller construct(~1700bp) and pcDNA5/FRT-backbone(~5000bp)]] | |
| + | |||
| + | To check the accuracy of the ligated pcDNA5/FRT-scFv-TEV-TMD-eYFP the construct was digested analytically using NheI and ApaI into the smaller construct exhibiting a bp size of approximately 1700 and into the pcDNA5/FRT backbone with approximately 5000bp. | ||
===== Expression and Localization of smaller construct===== | ===== Expression and Localization of smaller construct===== | ||
| Line 88: | Line 87: | ||
We carried out fluorescence microscopy with transiently as well as stably transfected cells. In both cases we detected yellow fluorescence which shows that the eYFP of the construct is expressed. Since eYFP is located at the C-terminal end of the sequence we can conclude that the whole construct inclusively scFv, transmembrane domain and TEV protease cleavage site is expressed in the cells. | We carried out fluorescence microscopy with transiently as well as stably transfected cells. In both cases we detected yellow fluorescence which shows that the eYFP of the construct is expressed. Since eYFP is located at the C-terminal end of the sequence we can conclude that the whole construct inclusively scFv, transmembrane domain and TEV protease cleavage site is expressed in the cells. | ||
| - | [[file:UP12_CHO_transientlytransfectedcells1.jpg|left|300px|thumb| | + | [[file:UP12_CHO_transientlytransfectedcells1.jpg|left|300px|thumb|'''Figure 9:''' eYFP fluorescence of the small construct in transient transfected CHO cells]] |
| - | [[file:UP12_CHO_stabletransfected_clon.jpg|center|300px|thumb| | + | [[file:UP12_CHO_stabletransfected_clon.jpg|center|300px|thumb|'''Figure 10:''' eYFP fluorescence of the small construct in stably transfected CHO cells]] |
<br> | <br> | ||
| Line 95: | Line 94: | ||
| - | [[file:UP12_CHO_transientlytransfectedcells_confocal_BF1.jpg|left|300px|thumb| | + | [[file:UP12_CHO_transientlytransfectedcells_confocal_BF1.jpg|left|300px|thumb|'''Figure 11:''' Brightfield image of CHO cell. Scale bar 10µm]] |
| - | [[file:UP12_CHO_transientlytransfectedcells_confocal_merge.jpg|center|300px|thumb| | + | [[file:UP12_CHO_transientlytransfectedcells_confocal_merge.jpg|center|300px|thumb|'''Figure 12:''' Overlay image of the small constract with scFV with eYFP. Scale bar 10µm]] |
<br> | <br> | ||
| Line 103: | Line 102: | ||
In order to detect the scFv in the cells we applied immunfluorescence. For this we used an antibody directed against the flag-tag included in the construct. This antibody was detected with an Alexa Fluor® coupled secondary anti-mouse antibody. The experiment was carried out with fixed cells and with fixed and permeabilized cells. Cells that were permeabilized were stained blue during the experiment. From this we can deduce that the scFv is expressed in the cells. The yellow fluorescence of the construct and the blue fluorescence of the detection antibody show co-localization in the overlay images. (see Figures) | In order to detect the scFv in the cells we applied immunfluorescence. For this we used an antibody directed against the flag-tag included in the construct. This antibody was detected with an Alexa Fluor® coupled secondary anti-mouse antibody. The experiment was carried out with fixed cells and with fixed and permeabilized cells. Cells that were permeabilized were stained blue during the experiment. From this we can deduce that the scFv is expressed in the cells. The yellow fluorescence of the construct and the blue fluorescence of the detection antibody show co-localization in the overlay images. (see Figures) | ||
<center><table><tr><td> | <center><table><tr><td> | ||
| - | [[file:UP12_immunfluorescence_eYFP.jpg|300px|thumb| | + | [[file:UP12_immunfluorescence_eYFP.jpg|300px|thumb|'''Figure 13:''' Fluorescence image of the eYFP attached to the small scFv construct]]</td><td> |
| - | [[file:UP12_immunfluorescence_CFP.jpg|300px|thumb| | + | [[file:UP12_immunfluorescence_CFP.jpg|300px|thumb|'''Figure 14:''' Immunfluorescence with Alexa Fluor® coupled secondary anti-mouse antibody directed against the flag-tag included in small scFv construct]]</td></tr></table></center> |
| - | [[file:UP12_immunfluorescence_merge.jpg|center|300px|thumb| | + | [[file:UP12_immunfluorescence_merge.jpg|center|300px|thumb|'''Figure 15:''' Overlay of the eYFP fluorescence image and the Alexa Fluor® coupled secondary anti-mouse antibodyC fluorescence image]] |
<br> | <br> | ||
| Line 113: | Line 112: | ||
===== FACS ===== | ===== FACS ===== | ||
| - | With regard to the mutation module we co-transfected cells with the smaller antibody construct and the modified AID with eGFP. These cells could successfully be sorted via FACS which again shows the expression of the | + | With regard to the mutation module we co-transfected cells with the smaller antibody construct with the anti-EGFR scFv and the modified AID with eGFP reporter. These cells could successfully be sorted via FACS which again shows the sufficient expression of the antibody construct with eYFP reporter. |
| - | + | ==== Transfection in HeLa and HEK AAV 293 cells ==== | |
| - | In order | + | In order to find out whether the transport of the small scFV antibody construct to the membrane is more efficient in other cell lines than CHO, we transiently transfected the small and the advanced antibody construct in HEK AAV 293 cells and HeLa cells. Both cell lines expressed the transfected antibodies but we were not able to detect a more efficient membrane presentation of antibody constructs. |
| - | [[file:UP12_HeLa_small_construct.jpg| | + | <center><table><tr><td> |
| - | < | + | [[file:UP12_HeLa_small_construct.jpg|left|300px|thumb|'''Figure 16:''' Localization of small scFV antibody constructs in the ER in transient transfected HeLa cells ]] |
| - | [[file: | + | </td><td> |
| + | [[file:UP12_Nano_HeLa.jpg|center|300px|thumb|'''Figure 17:''' Localization of big nanobody constructs in the ER in transient transfected HeLa cells ]] | ||
| + | </td></tr></table></center> | ||
| + | <center><table><tr><td> | ||
| + | [[file: UP12_Image_scFv_HEK.jpg|left|300px|thumb|'''Figure 18:''' Localization of small scFV antibody constructs in the ER in transient transfected HEK AAV 293 cells ]] | ||
| + | </td><td> | ||
| + | [[file: UP12_Nano_HEK.jpg|center|300px|thumb|'''Figure 19:''' Localization of big nanobody construct in the ER in transient transfected HEK AAV 293 cells ]] | ||
| + | </td></tr></table></center> | ||
<br> | <br> | ||
| Line 128: | Line 134: | ||
The advanced antibody construct was designed by us de novo on the basis of sequence information provided by Genebank and UniProt files and one already existing Biobrick part. The construct consists of two major building blocks represented by the actual antibody unit and the switchable membrane anchoring region. The antibody unit is represented by the signal peptide, the anti-GFP Nanobody and the Fc region. TEV protease recognition site, 2 LoxP sites, the B-cell receptor transmembrane domain and the mCherry reporter display the switchable membrane anchoring region. The signal peptide was taken from a human immunoglobulin G kappa variable chain I (UniProt: P01601) and functions as translocation signal to the cell membrane. As antibody component for this construct we used a single-chain VHH antibody domain, also called nanobody. The anti- green fluorescent protein (GFP)-nanobody was developed for specific binding activity to GFP. (see Introduction) (Kubala et al; 2010) Our advanced construct possesses a special feature that allows the replacement of the nanobody with another antigen binding domain. The nanobody is framed by two specific restriction enzyme recognition sites, SacI on the N-terminal end and KasI at the C-terminal end, which will allow the easy exchange with another favored and suitable sequence. The nanobody is fused to a human immunoglobulin gamma 1 heavy chain constant region (Fc) (UniProt: P01857) which enables the direct interaction of the antibody unit with the Fc receptor and complement proteins. Similar to the smaller construct a TEV protease cleavage site was integrated between the Fc region and the transmembrane domain permitting as well the shift from surface presentation to secretion on protein level (see smaller construct). For an unimpeded activity of the TEV protease two amino acids at the N-terminal end were added as spacer. Additionally to this switch on protein level, the whole membrane anchoring region is flanked by two LoxP sites introducing a non-reversible genetically switch to our system. It allows the modification from surface presentation to secretion on the DNA level. Both LoxP sites are oriented in the same direction leading to the elimination of the enclosed region by cre recombinase activity. We choose two LoxP site sequences of 34 base pairs (ATAACTTCGTATA GCATACAT TATACGAAGTTAT) for recognition by cre recombinase. The helical single-span transmembrane domain sequence of the B-cell receptor was taken from the registry (BBa_K157010) and modified by shortening of the linker sequence to three amino acids at the C-terminal end. Expression of the advanced antibody construct and its cellular localization can be monitored by the mCherry signal with an excitation at 587 nm and an emission at 610 nm. | The advanced antibody construct was designed by us de novo on the basis of sequence information provided by Genebank and UniProt files and one already existing Biobrick part. The construct consists of two major building blocks represented by the actual antibody unit and the switchable membrane anchoring region. The antibody unit is represented by the signal peptide, the anti-GFP Nanobody and the Fc region. TEV protease recognition site, 2 LoxP sites, the B-cell receptor transmembrane domain and the mCherry reporter display the switchable membrane anchoring region. The signal peptide was taken from a human immunoglobulin G kappa variable chain I (UniProt: P01601) and functions as translocation signal to the cell membrane. As antibody component for this construct we used a single-chain VHH antibody domain, also called nanobody. The anti- green fluorescent protein (GFP)-nanobody was developed for specific binding activity to GFP. (see Introduction) (Kubala et al; 2010) Our advanced construct possesses a special feature that allows the replacement of the nanobody with another antigen binding domain. The nanobody is framed by two specific restriction enzyme recognition sites, SacI on the N-terminal end and KasI at the C-terminal end, which will allow the easy exchange with another favored and suitable sequence. The nanobody is fused to a human immunoglobulin gamma 1 heavy chain constant region (Fc) (UniProt: P01857) which enables the direct interaction of the antibody unit with the Fc receptor and complement proteins. Similar to the smaller construct a TEV protease cleavage site was integrated between the Fc region and the transmembrane domain permitting as well the shift from surface presentation to secretion on protein level (see smaller construct). For an unimpeded activity of the TEV protease two amino acids at the N-terminal end were added as spacer. Additionally to this switch on protein level, the whole membrane anchoring region is flanked by two LoxP sites introducing a non-reversible genetically switch to our system. It allows the modification from surface presentation to secretion on the DNA level. Both LoxP sites are oriented in the same direction leading to the elimination of the enclosed region by cre recombinase activity. We choose two LoxP site sequences of 34 base pairs (ATAACTTCGTATA GCATACAT TATACGAAGTTAT) for recognition by cre recombinase. The helical single-span transmembrane domain sequence of the B-cell receptor was taken from the registry (BBa_K157010) and modified by shortening of the linker sequence to three amino acids at the C-terminal end. Expression of the advanced antibody construct and its cellular localization can be monitored by the mCherry signal with an excitation at 587 nm and an emission at 610 nm. | ||
<br> | <br> | ||
| - | [[file:UP12_BBa_K929107.png| | + | [[file:UP12_BBa_K929107.png|left|600px|thumb|'''Figure 20:''' Advanced antibody construct cloned into pSB1C3 (BBa_K929107)]] |
| - | + | [[file:UP12_nanobodypcdna5frtcloning.png|center|150px|thumb|'''Figure 21:''' Test digestion of advanced construct with NheI and ApaI; 1: marker, 2: advanced construct(~2200bp) and pcDNA5/FRT-backbone(~5000bp)]] | |
| + | |||
| + | To verify the accuracy of the ligated pcDNA5/FRT-advanced antibody construct an analytical digestion using NheI and ApaI was perfomred.The advanced antibody construct appears at a bp size of approximately 2200 and the pcDNA5/FRT backbone at approximately 5000bp. | ||
===== Expression and Localization of advanced construct===== | ===== Expression and Localization of advanced construct===== | ||
| Line 141: | Line 149: | ||
| - | [[file:UP12_CHO_stabletransfected_Big_Konstruct.jpg|center| | + | [[file:UP12_CHO_stabletransfected_Big_Konstruct.jpg|center|500px|thumb|'''Figure 22:''' Membrane localization of advanced nanobody construct in stably transfected CHO cells ]] |
<br> | <br> | ||
| - | [[file:UP12_CHO_stabletransfected_Big_Konstruct_no membrane loacalisation.jpg|center| | + | <center><table><tr><td> |
| - | [[file:UP12_ER_localisation.jpg|center| | + | [[file:UP12_CHO_stabletransfected_Big_Konstruct_no membrane loacalisation.jpg|center|350px|thumb|'''Figure 23:''' Advanced nanobody construct with seemingly no membrane localization in stably transfected CHO cells]]</td><td> |
| + | [[file:UP12_ER_localisation.jpg|center|350px|thumb|'''Figure 24:''' Advanced nanobody construct with localisation in ER in stably transfected CHO cells]]</td></tr></table></center> | ||
<br> | <br> | ||
| Line 154: | Line 163: | ||
From this we can conclude that the Cre Recombinase was expressed and that the antibody was secreted after the removal of the transmembrane domain and the mCherry. | From this we can conclude that the Cre Recombinase was expressed and that the antibody was secreted after the removal of the transmembrane domain and the mCherry. | ||
| - | [[file: | + | [[file:UP12_Cre_Recombinase_Blot_ohneBildunterschrift.png|center|300px|thumb|'''Figure 25:''' Western Blot of purified Nanobody-Fc from supernatant of cells co-transfected with Cre recombinase and advanced construct. Purification was carried out with magnetic beads. Correct band can be seen at 41kDa]]<br> |
===== FACS ===== | ===== FACS ===== | ||
| Line 163: | Line 172: | ||
In order fo find out whether the transport of the antibody constructs to the membrane is more efficient in other cell lines than CHO we transiently transfected the small and the advanced construct in AAV 293 cells and HeLa cells. Both cell lines expressed the transfected antibodies but we were not able to detect a more efficient membrane presentation of antibody constructs. | In order fo find out whether the transport of the antibody constructs to the membrane is more efficient in other cell lines than CHO we transiently transfected the small and the advanced construct in AAV 293 cells and HeLa cells. Both cell lines expressed the transfected antibodies but we were not able to detect a more efficient membrane presentation of antibody constructs. | ||
| + | |||
| + | ==== Exchange of Signal Peptide and Transmembrane Domain==== | ||
| + | |||
| + | In order to achieve the membrane localization of our small antibody construct we exchanged the former signal peptide and the transmembrane domain. Both sequences were taken from the human receptor for advanced glycation end products (RAGE) (UniProt: Q15109). These RAGE proteins are nonenzymatically glycosylated proteins which accumulate in vascular tissue at aging processes and at an accelerated rate in diabetes. Its functional unit of signal peptide and transmembrane domain was successfully applied in CHO cells where it enabled the translocation to and integration into the outer membrane. | ||
| + | <br> | ||
| + | The signal peptide is composed of 22 amino acids (aa) and the helical transmembrane domain, originally occurring with 21 aa, was extended on each side by three aa of its flanking extracellular and cytoplasmic regions. | ||
| + | For the purpose of controlling the functionality of the new transmembrane domain we also equipped our advanced antibody construct with the RAGE sequence. Its original signal peptide of a human immunoglobulin G kappa variable chain I (UniProt: P01601) was maintained. | ||
=== Discussion === | === Discussion === | ||
| - | ... | + | We successfully designed two antibody constructs and transfected them into CHO cells. Both the small and the advanced antibody construct are expressed in full length as we have demonstrated with fluorescence and confocal microscopy, immunofluorescence, Western Blot and FACS. Although we have seen a membrane localization of the advanced antibody construct we were not able to generate cells that transported enough antibody molecules to their surface for our detection methods to work. We were able to detect the single chain variable domain fragment (scFV425) via the fluorescence of the eYFP and via immunofluorescence. This experiment showed the expression of the construct but since only permeabilized cells were stained with the secondary antibody we concluded that the protein is located inside the cells and is not sufficiently integrated into the outer cell membrane. This was confirmed with confocal microscopy where a strong localization of the scFv construct in the endoplasmatic reticulum could be detected. |
| + | |||
| + | The advanced antibody construct was also successfully expressed in CHO cells but was not always detectable in the membrane. We could not adjust the conditions or circumstances which lead to the successful transport of the antibody molecules to the membrane. | ||
| + | |||
| + | As a proof of the functionality of our cre/LoxP system we were able to show that the switch from presenting antibodies to secreting antibodies into the supernatant can be induced by the cre recombinase. This is a very important step in the establishment of our antibody generation system. | ||
| + | |||
| + | Since the transport and presentation of the antibody constructs shows the problematic of uncertain membrane integration in CHO cells as well as in HEK293 and HeLa cells we decided to modify our small and our advanced constructs membrane domain and signal sequence. The antibody constructs will then contain another signal peptide as well as another transmembrane domain. As new signal peptide and transmembrane domain we chose sequences from human receptor for advanced glycation end products (RAGE). Both constructs were successfully cloned and transfected into CHO cells. Currently we are working to gain experimental data. | ||
| + | |||
=== References === | === References === | ||
* de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb Cell Fact. 2011 Jun 9;10:44. Review. [http://www.ncbi.nlm.nih.gov/pubmed?term=21658216 PubMed PMID: 21658216.] | * de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb Cell Fact. 2011 Jun 9;10:44. Review. [http://www.ncbi.nlm.nih.gov/pubmed?term=21658216 PubMed PMID: 21658216.] | ||
Latest revision as of 18:41, 26 October 2012
Contents
|
Antibody Module
Abstract
The function of the antibody module is to design and test antibody constructs which operate as sequence substrate for the AID, our mutating module. The antibody constructs were stably and transiently transfected into CHO cells. The antigen binding domains of the construct can directly interact with the specific antigen on the surface of the virus construct, the work object of our selection module. This interaction can lead to selective collection of a mutated and thus optimized antibody.
Introduction
Aim of the Antibody Module
The idea of the antibody module is to create antibody constructs which will specifically be mutated and optimized by the AID (activation-induced cytidine deaminase). Our two antibody constructs allow an easy handling and a straight forward integration into CHO cells. The use of Flp-In system realizes time saving incorporation of antibody constructs into the CHO genome. A switch from surface presentation to secretion of the antibodies can be enabled by TEV protease and cre recombinase and permits a simple harvest of selected antibodies.
A Short Review on Antibodies
Antibodies are highly specific targeting reagents and are the key defense system for recognizing pathogens and toxins. Antibodies are Y shaped multi domain proteins with two antigen binding sites displaying the Fab fragment and one effector domain represented by the Fc domain. (Holliger, Hudson; 2005) The Fab fragment is composed of two antigen recognition sites with one variable light (VL) and constant light (VC) chain and one variable heavy (VH) and constant heavy (CH) chain each which allow the specific and affine binding of antigens. By interacting with the complement and the Fc receptor, the Fc domain is able to mediate cytotoxic effector functions. (Holliger, Hudson; 2005)
Single Chain Fragment Variable Domain
A single chain fragment variable is a small unit of immunoglobulin and consists of the variable heavy (VH) and the variable light (VL) domains which are joined together by a flexible peptide linker. (Alitheen and Hamid et al.; 2012) The scFv is a very popular format that shows a high antigen binding capacity and can be easily expressed.
Nanobody
A specific form of immunoglobulin is the single domain antibody fragment, called by its inventor Ablynx - nanobody. Nanobodies are derived from camelid antibodies that possess a single N-terminal domain, the VHH domain. The VHH domain is solely sufficient for a strong antigen binding and does not require a domain fusion. (Harmsen, Haard; 2007) Thus, nanobodies have extremely small dimensions and show an elevated stability which both leads to the ability of recognizing epitopes that are not accessible for conventional antibody formats. (de Marco; 2011)
Generating the Antibody Constructs
Two antibody constructs with different constitutions were generated by us to diversify our system and to guarantee a broadened scientific approach. They do also allow a directed troubleshoot of our concept.
Smaller Antibody Construct
We designed the smaller antibody construct out of already existing parts and BioBricks and added further functional units by assembly PCR. The smaller construct is built up from a single chain fragment (anti-EGRF scFv425) with a signal peptide for membrane translocation (BBa_K157001) on its N-terminal end, the transmembrane domain (BBa_K157010) with a flanking TEV recognition site and the eYFP reporter (BBa_E0030) at its C-terminal end.
The scFv425 is of murine origin and was derived against the human epidermal growth factor receptor domain 3. It was originally described in context of a publication dealing with immunotherapy of pancreatic carcinoma cells which consequently express the EGFR. The scFv was used as mediator of carcinoma cells and an endotoxin, thus showing the high relevance of the single chain fragment for medical purposes. (Bruell et al; 2005)
Advanced Antibody Construct
The advanced antibody construct was designed de novo via gene synthesis. It consists of two major building blocks represented by the actual antibody unit and the switchable membrane anchoring region. Both elements guarantee the eligibility and easy handling of the construct in our CHO cell Flp-In expression system and are codon optimized.
The central part is the antibody cassette consisting of a replaceable anti-GFP nanobody and an IgG1 stem Fc domain (GenBank: J00228.1). A nanobody-Fc fusion protein combines the advantage of smaller dimensions with the ability to directly interact with the Fc receptor and complement proteins. The green fluorescent protein (GFP)-nanobody is a single-chain VHH antibody domain developed for specific binding activity to GFP and shows a Kd value of 1.4 nM. Its CDR3 loop is very short and has significantly less contacts with the GFP ligand compared to other nanobodies. Furthermore, the shortness of this CDR3 loop leads to the exposure of the framework 2 region, which has a major contribution to the binding with GFP. (Kubala et al; 2010) The nanobody is framed by two specific restriction enzyme recognition sites on each terminal end allowing the easy exchange of this part with another suitable sequence. The membrane anchoring region (modified BBa_K157010) is flanked by two LoxP sites that introduce a genetical cut to our system which will allow the modification from surface presentation to secretion of the antibody construct by activity of cre recombinase (Araki et al; 1997) in our CHO cells. The cre recombinase is a trysosine recombinase enzyme which catalyzes reciprocal site-specific recombination between two specific 34-bp sites called loxP. The cre/loxP recombination system was derived from bacteriophage P1 and is frequently used in genetic manipulation. (Araki et al; 1997)
Additionallly we integrated a switch on the protein level which is represented by the TEV protease cleavage site permitting the shift to a secretory antibody production. Expression of the antibody construct and its cellular localization can be monitored by the mCherry signal.
CHO Cells and Flp-In System
Chinese hamster ovary cells (CHO) are an immortal mammalian cell line which is frequently used for industrial recombinant protein expressions. Problems when working with mammalian cells are the varying transfection rate and the transitory nature of transient transfected plasmids. To overcome this problem we are working with a CHO expression cell line for the stable transfection of our construct of interest by using Flp recombinase-mediated integration.
Flp-In System
The FRT/Flp system (LifeTechnologies) is based on homologous recombination initiated by the enzyme flippase (Flp) at a specific FRT site in the genome. CHO cell containing a randomly but stably integrated FRT sequence can be selected by antibiotic resistance against Zeocin. Another FRT sequence is located on the shuttle plasmid (pcDNA/5FRT) that contains the gene of interest and a second antibiotic resistance against Hygromycin. The system is accomplished by the third plasmid (pOG44) carrying the recombinase flippase (Flp) which mediates the recombination of both FRT sequences leading to an exchange of Zeocin to Hygromycin resistance.
Both of our antibody constructs were also transfected transiently to get a first impression of the expression level, the cellular localization and the compatibility with the CHO cell metabolism.
Working with Other Cell Lines
In order to get a better insight into our system and its way of function we also transiently transfected HeLa and HEK293 cells with our constructs. HeLa cells are the oldest and most commonly used immortal human cell line and were derived in 1951 from cervical cancers cells of a patient called Henrietta Lacks. The other cell line we are using is HEK293 cells which are immortal human embryonic kidney cells.
Results
CHO cells
Cell Duplication Rate
The duplication rate of CHO Flp-In cells was estimated from cell countings, seeding numbers and observation of confluency. Cells that were seeded at a density of 8*10^5 in 75cm² culture flasks reached confluency after 48h. We concluded that cells duplicate every 10 to 15h.
Sensitivity to Hygromycin for Flp-In System
After 48 hours of co-transfection of CHO cells with pOG 44 and the Flp-in expression construct we selected stable transfected cells by using Hygromycin B. Before we started with transfection we determined the optimal Hygromycin concentration by testing the sensitivity of the CHO Flp-In cells. For this purpose we applied different Hygromycin concentrations over a period of 6 days and determined cell numbers by counting cells every 24h. Since Hygromycin only affects growing cells we looked for the concentration at which cell growth was prevented. According to the data shown in Figure 6 we decided to carry out the selection with a Hygromycin concentration of 550 µg/ml.
Smaller Antibody Construct
Cloning the Smaller Construct
The sequence from the human EGFR (ErbB-1) signal peptide was taken from the already existing BioBrick BBa_K157001 and was fused N-terminally to the scFv 425 against the epidermal growth factor receptor (EGFR) domain 3. The scFv425 has a shortened N-terminal FLAG-tag sequence of five amino acids (DYKDE) that allows us a handy purification and simplifies its detection. A TEV protease cleavage site was integrated between the scFv and the transmembrane domain by primer extension and permits the shift from surface presentation to secretion on protein level. The TEV protease recognition site ENLYFQG was used here and represents the most commonly used aa sequence for recognition by the 27kDA catalytic domain of Nuclear Inclusion a (NIa) protein encoded by the tobacco etch virus (TEV). For an unimpeded activity of the TEV protease a three amino acid linker at the N-terminal end and a two amino acid linker at the C-terminal end were added to the sequence. The helical single-span transmembrane domain of the B-cell receptor (BBa_K157010) was modified in terms of the shortening of the linker sequence to three amino acids at the C-terminal end. Expression of the scFv fusion protein and its cellular localization can be monitored by the enhanced YFP (BBa_E0030) signal with an excitation at 514 nm and an emission at 527 nm.
To check the accuracy of the ligated pcDNA5/FRT-scFv-TEV-TMD-eYFP the construct was digested analytically using NheI and ApaI into the smaller construct exhibiting a bp size of approximately 1700 and into the pcDNA5/FRT backbone with approximately 5000bp.
Expression and Localization of smaller construct
Expression of the smaller antibody construct with scFv in CHO Flp-In cells was proven with different methods such as fluorescence and confocal microscopy, immunfluorescence and FACS.
Microscopy
We carried out fluorescence microscopy with transiently as well as stably transfected cells. In both cases we detected yellow fluorescence which shows that the eYFP of the construct is expressed. Since eYFP is located at the C-terminal end of the sequence we can conclude that the whole construct inclusively scFv, transmembrane domain and TEV protease cleavage site is expressed in the cells.
Yellow fluorescence could also be shown with confocal microscopy. Here we also observed that the fluorescence seems to be centered around the nucleus and is spread in the endoplasmatic reticulum (ER).
Immunfluorescence
In order to detect the scFv in the cells we applied immunfluorescence. For this we used an antibody directed against the flag-tag included in the construct. This antibody was detected with an Alexa Fluor® coupled secondary anti-mouse antibody. The experiment was carried out with fixed cells and with fixed and permeabilized cells. Cells that were permeabilized were stained blue during the experiment. From this we can deduce that the scFv is expressed in the cells. The yellow fluorescence of the construct and the blue fluorescence of the detection antibody show co-localization in the overlay images. (see Figures)
Cells that were fixed and not permeabilized were expected to be stained on the outside since the antibody construct is supposed to be localized in the membrane. In the microscope we were not able to detect any blue fluorescence on the surface of the cells which leads us to the conclusion that there was not enough antibody transported to the surface for detection.
FACS
With regard to the mutation module we co-transfected cells with the smaller antibody construct with the anti-EGFR scFv and the modified AID with eGFP reporter. These cells could successfully be sorted via FACS which again shows the sufficient expression of the antibody construct with eYFP reporter.
Transfection in HeLa and HEK AAV 293 cells
In order to find out whether the transport of the small scFV antibody construct to the membrane is more efficient in other cell lines than CHO, we transiently transfected the small and the advanced antibody construct in HEK AAV 293 cells and HeLa cells. Both cell lines expressed the transfected antibodies but we were not able to detect a more efficient membrane presentation of antibody constructs.
Advanced Antibody Construct
Gene synthesis
The advanced antibody construct was designed by us de novo on the basis of sequence information provided by Genebank and UniProt files and one already existing Biobrick part. The construct consists of two major building blocks represented by the actual antibody unit and the switchable membrane anchoring region. The antibody unit is represented by the signal peptide, the anti-GFP Nanobody and the Fc region. TEV protease recognition site, 2 LoxP sites, the B-cell receptor transmembrane domain and the mCherry reporter display the switchable membrane anchoring region. The signal peptide was taken from a human immunoglobulin G kappa variable chain I (UniProt: P01601) and functions as translocation signal to the cell membrane. As antibody component for this construct we used a single-chain VHH antibody domain, also called nanobody. The anti- green fluorescent protein (GFP)-nanobody was developed for specific binding activity to GFP. (see Introduction) (Kubala et al; 2010) Our advanced construct possesses a special feature that allows the replacement of the nanobody with another antigen binding domain. The nanobody is framed by two specific restriction enzyme recognition sites, SacI on the N-terminal end and KasI at the C-terminal end, which will allow the easy exchange with another favored and suitable sequence. The nanobody is fused to a human immunoglobulin gamma 1 heavy chain constant region (Fc) (UniProt: P01857) which enables the direct interaction of the antibody unit with the Fc receptor and complement proteins. Similar to the smaller construct a TEV protease cleavage site was integrated between the Fc region and the transmembrane domain permitting as well the shift from surface presentation to secretion on protein level (see smaller construct). For an unimpeded activity of the TEV protease two amino acids at the N-terminal end were added as spacer. Additionally to this switch on protein level, the whole membrane anchoring region is flanked by two LoxP sites introducing a non-reversible genetically switch to our system. It allows the modification from surface presentation to secretion on the DNA level. Both LoxP sites are oriented in the same direction leading to the elimination of the enclosed region by cre recombinase activity. We choose two LoxP site sequences of 34 base pairs (ATAACTTCGTATA GCATACAT TATACGAAGTTAT) for recognition by cre recombinase. The helical single-span transmembrane domain sequence of the B-cell receptor was taken from the registry (BBa_K157010) and modified by shortening of the linker sequence to three amino acids at the C-terminal end. Expression of the advanced antibody construct and its cellular localization can be monitored by the mCherry signal with an excitation at 587 nm and an emission at 610 nm.
To verify the accuracy of the ligated pcDNA5/FRT-advanced antibody construct an analytical digestion using NheI and ApaI was perfomred.The advanced antibody construct appears at a bp size of approximately 2200 and the pcDNA5/FRT backbone at approximately 5000bp.
Expression and Localization of advanced construct
Expression of the advanced antibody construct with Nanobody in CHO Flp-In cells was proved with different methods such as fluorescence microscopy, Western Blot and FACS.
Microscopy
We carried out fluorescence microscopy with transiently as well as stably transfected cells. In both cases we detected red fluorescence which shows that the mCherry of the construct is expressed. Since mCherry is located at the c-terminal end of the sequence we can conclude that the whole construct including the Nanobody, Fc-part, transmembrane domain, LoxP sites and the TEV protease cleavage site is expressed in the cells.
During our experiments we found that at some times membrane localisation of the advanced construct can be observed. At other times and under different conditions however the constructs seem to be localized in the ER.
Cre Recombinase
In order to show the switch from presenting the antibodies to sectreting them into the supernatant we applied SDS-PAGE and Western Blot. CHO cells were co-transfected with the advanced antibody construct and the Cre Recombinase and the supernatant was collected after 3 days of incubation. In order to test the functionality of cre recombinase we incubated the supernatant with magnetic beads coupled to GFP via streptavidin and biotin. Afterwards, we did a Western blot to check if the anti-GFP nanobody is expressed and can bind to GFP. In the Western Blot we detected the nanobody-Fc construct with an anti-human Fc antibody. We got a band at 41kDa which correlates with the size of our Fc part and the nanobody. The negative controls (beads without GFP, Medium containing FBS and non transfected CHO cells) did not show any bands in the blot. From this we can conclude that the Cre Recombinase was expressed and that the antibody was secreted after the removal of the transmembrane domain and the mCherry.
FACS
With regard to the mutation module we co-transfected cells with the advanced antibody construct and the modified AID with eGFP. These cells could successfully be sorted via FACS which again shows the expression of the mCherry of the advanced construct.
Transfection in HeLa and HEK cells
In order fo find out whether the transport of the antibody constructs to the membrane is more efficient in other cell lines than CHO we transiently transfected the small and the advanced construct in AAV 293 cells and HeLa cells. Both cell lines expressed the transfected antibodies but we were not able to detect a more efficient membrane presentation of antibody constructs.
Exchange of Signal Peptide and Transmembrane Domain
In order to achieve the membrane localization of our small antibody construct we exchanged the former signal peptide and the transmembrane domain. Both sequences were taken from the human receptor for advanced glycation end products (RAGE) (UniProt: Q15109). These RAGE proteins are nonenzymatically glycosylated proteins which accumulate in vascular tissue at aging processes and at an accelerated rate in diabetes. Its functional unit of signal peptide and transmembrane domain was successfully applied in CHO cells where it enabled the translocation to and integration into the outer membrane.
The signal peptide is composed of 22 amino acids (aa) and the helical transmembrane domain, originally occurring with 21 aa, was extended on each side by three aa of its flanking extracellular and cytoplasmic regions.
For the purpose of controlling the functionality of the new transmembrane domain we also equipped our advanced antibody construct with the RAGE sequence. Its original signal peptide of a human immunoglobulin G kappa variable chain I (UniProt: P01601) was maintained.
Discussion
We successfully designed two antibody constructs and transfected them into CHO cells. Both the small and the advanced antibody construct are expressed in full length as we have demonstrated with fluorescence and confocal microscopy, immunofluorescence, Western Blot and FACS. Although we have seen a membrane localization of the advanced antibody construct we were not able to generate cells that transported enough antibody molecules to their surface for our detection methods to work. We were able to detect the single chain variable domain fragment (scFV425) via the fluorescence of the eYFP and via immunofluorescence. This experiment showed the expression of the construct but since only permeabilized cells were stained with the secondary antibody we concluded that the protein is located inside the cells and is not sufficiently integrated into the outer cell membrane. This was confirmed with confocal microscopy where a strong localization of the scFv construct in the endoplasmatic reticulum could be detected.
The advanced antibody construct was also successfully expressed in CHO cells but was not always detectable in the membrane. We could not adjust the conditions or circumstances which lead to the successful transport of the antibody molecules to the membrane.
As a proof of the functionality of our cre/LoxP system we were able to show that the switch from presenting antibodies to secreting antibodies into the supernatant can be induced by the cre recombinase. This is a very important step in the establishment of our antibody generation system.
Since the transport and presentation of the antibody constructs shows the problematic of uncertain membrane integration in CHO cells as well as in HEK293 and HeLa cells we decided to modify our small and our advanced constructs membrane domain and signal sequence. The antibody constructs will then contain another signal peptide as well as another transmembrane domain. As new signal peptide and transmembrane domain we chose sequences from human receptor for advanced glycation end products (RAGE). Both constructs were successfully cloned and transfected into CHO cells. Currently we are working to gain experimental data.
References
- de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb Cell Fact. 2011 Jun 9;10:44. Review. [http://www.ncbi.nlm.nih.gov/pubmed?term=21658216 PubMed PMID: 21658216.]
- Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol. 2007 Nov;77(1):13-22. Epub 2007 Aug 18. Review. [http://www.ncbi.nlm.nih.gov/pubmed?term=17704915 PubMed PMID: 17704915.]
- Peterson E, Owens SM, Henry RL. Monoclonal antibody form and function: manufacturing the right antibodies for treating drug abuse. AAPS J. 2006 May 26;8(2):E383-90. Review. [http://www.ncbi.nlm.nih.gov/pubmed?term=16796389 PubMed PMID: 16796389.]
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005 Sep;23(9):1126-36. Review. [http://www.ncbi.nlm.nih.gov/pubmed?term=16151406 PubMed PMID: 16151406.]
- Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NB, Hamid M. scFv antibody: principles and clinical application. Clin Dev Immunol. 2012;2012:980250. Epub 2012 Mar 15. [http://www.ncbi.nlm.nih.gov/pubmed?term=22474489 PubMed PMID: 22474489.]
- Bruell D, Bruns CJ, Yezhelyev M, Huhn M, Müller J, Ischenko I, Fischer R, Finnern R, Jauch KW, Barth S. Recombinant anti-EGFR immunotoxin 425(scFv)-ETA' demonstrates anti-tumor activity against disseminated human pancreatic cancer in nude mice. Int J Mol Med. 2005 Feb;15(2):305-13. [http://www.ncbi.nlm.nih.gov/pubmed?term=15647848 PubMed PMID: 15647848.]
- Invitrogen (2011) Flp-In System Manual. Publication Part Number 25-0366
 "
"