Team:University College London/LabBook/Week16
From 2012.igem.org
(→16-1) |
(→16-1) |
||
| (22 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<div class="experimentContent"></html> | <div class="experimentContent"></html> | ||
==16-1== | ==16-1== | ||
| + | <html><div class="protocol protocol-Generic">DNAse assay</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/DNAse_assay}}<html></div></html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | {| class = "wikitable" | ||
| + | |- | ||
| + | !Sample/time !! Colony diameter/mm !! Halo diameter/mm !! Absorbance at 600 OD | ||
| + | |- | ||
| + | |1 (12.30 noon) || 0 || 0 || 0 | ||
| + | |- | ||
| + | |2 (16.30pm) || 0 || 0 || 0 | ||
| + | |- | ||
| + | |3 (19.30 pm) || 0 || 0 || 0 | ||
| + | |- | ||
| + | |4 (22.30 pm) || 0.5 || 1 || 0.002 | ||
| + | |- | ||
| + | |5 (01.30 am) || 1 || 3.5 || 0.182 | ||
| + | |- | ||
| + | |6 (04.30 am) || 1.5 || 7 || 0.238 | ||
| + | |- | ||
| + | |7 (07.30 am) || 2 || 9 || 0.297 | ||
| + | |- | ||
| + | |8 (10.30am) || 2.5 || 11 || 0.518 | ||
| + | |- | ||
| + | |9 (12.30noon) || 3 || 12.5 || 0.701 | ||
| + | |- | ||
| + | |10 (2.30pm) || 3.5 || 14 || 0.811 | ||
| + | |- | ||
| + | |11(4.30pm) || 4 || 15 || 0.906 | ||
| + | |} | ||
| + | |||
| + | '''Conclusion:''' | ||
| + | From the table above we concluded that there is a positive correlation between the diametr of the colony and the diameter of the halo and both increase over time. This proves that our ligation of T7+nuclease was successful. | ||
| + | |||
<html></div> | <html></div> | ||
<div class="experiment"></div> | <div class="experiment"></div> | ||
| Line 12: | Line 46: | ||
==Monday (24.09.12) == | ==Monday (24.09.12) == | ||
<html><div class="protocol protocol-Generic">Congo Red Assay</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Congo_Red_Assay}}<html></div></html> | <html><div class="protocol protocol-Generic">Congo Red Assay</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Congo_Red_Assay}}<html></div></html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ Congo Red Agar Assay | ||
| + | ! Control (W3110) !! BBa_K729017 | ||
| + | |- | ||
| + | | [[File:UniversityCollegeLondon_Curli_Congo_ Red_Control.jpg|475px]] || [[File:UniversityCollegeLondon_Curli_Congo_ Red_K729017.jpg|475px]] | ||
| + | |} | ||
| + | |||
| + | From the plates above we can see that our BL21 control that does not have T7+curli insert and does not appear red. However, BL21 that has T7+curli insert appears red. BL21 with a constitutive promoter also shows uptake of dye and shoiws red colonies. | ||
| + | |||
| + | |||
| + | '''Conclusion:''' | ||
| + | We conclude successful expression of the curli operon as indicated by the red pigmented colonies. Ability to uptake congo red dye resides in the amyloid structure of the appendages and ionic interactions. | ||
| + | |||
<html> | <html> | ||
</div><div class="experiment"></div><img src="https://static.igem.org/mediawiki/2012/3/32/Ucl2012-labbook-graph16-3.png" /><div class="experimentContent"> | </div><div class="experiment"></div><img src="https://static.igem.org/mediawiki/2012/3/32/Ucl2012-labbook-graph16-3.png" /><div class="experimentContent"> | ||
Latest revision as of 03:42, 27 September 2012



16-1
1. Prepare 11-16 plates (DNAse - Agar + CMP)
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Apply HCl to the first plate before putting in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings every two hours.
6. When the reading is taken, observe the following: a. Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours) b. Diameter of the halo that is achieved once HCL is applied c. OD from a) d. Estimate of the depth of the colony on the agar plate
Results:
| Sample/time | Colony diameter/mm | Halo diameter/mm | Absorbance at 600 OD |
|---|---|---|---|
| 1 (12.30 noon) | 0 | 0 | 0 |
| 2 (16.30pm) | 0 | 0 | 0 |
| 3 (19.30 pm) | 0 | 0 | 0 |
| 4 (22.30 pm) | 0.5 | 1 | 0.002 |
| 5 (01.30 am) | 1 | 3.5 | 0.182 |
| 6 (04.30 am) | 1.5 | 7 | 0.238 |
| 7 (07.30 am) | 2 | 9 | 0.297 |
| 8 (10.30am) | 2.5 | 11 | 0.518 |
| 9 (12.30noon) | 3 | 12.5 | 0.701 |
| 10 (2.30pm) | 3.5 | 14 | 0.811 |
| 11(4.30pm) | 4 | 15 | 0.906 |
Conclusion: From the table above we concluded that there is a positive correlation between the diametr of the colony and the diameter of the halo and both increase over time. This proves that our ligation of T7+nuclease was successful.

Monday (24.09.12)
Congo red binding assay, Preparation of Congo red supplemented agar:
20uL.ml-1 Congo Red Dye 10ug.ml-1 Coomassie brilliant blue dye G-250 LB Agar (No salt)
Plates subsequently streaked. Uptake of red pigment indicative of amyloid fiber expression.
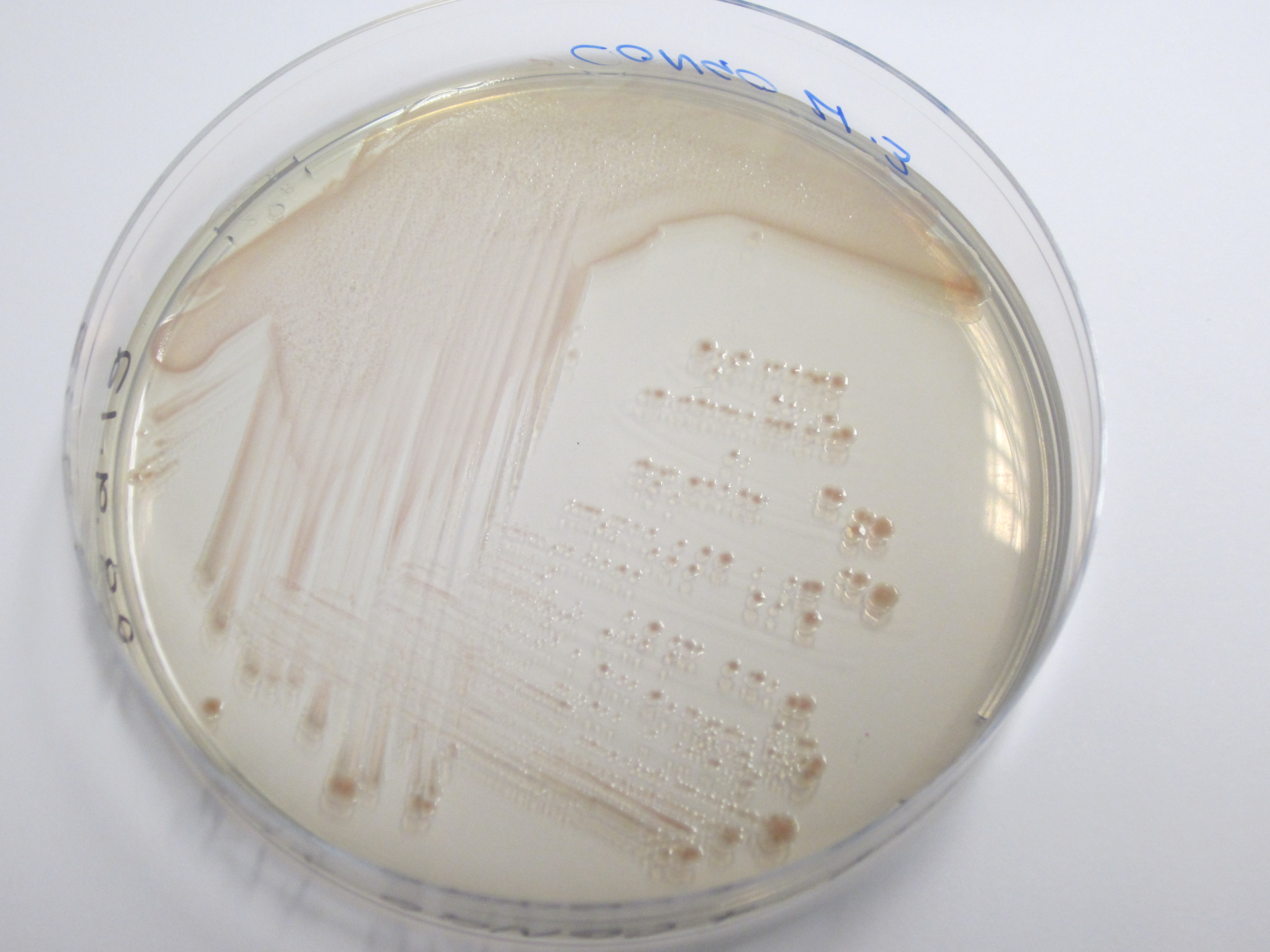
Results:
| Control (W3110) | BBa_K729017 |
|---|---|
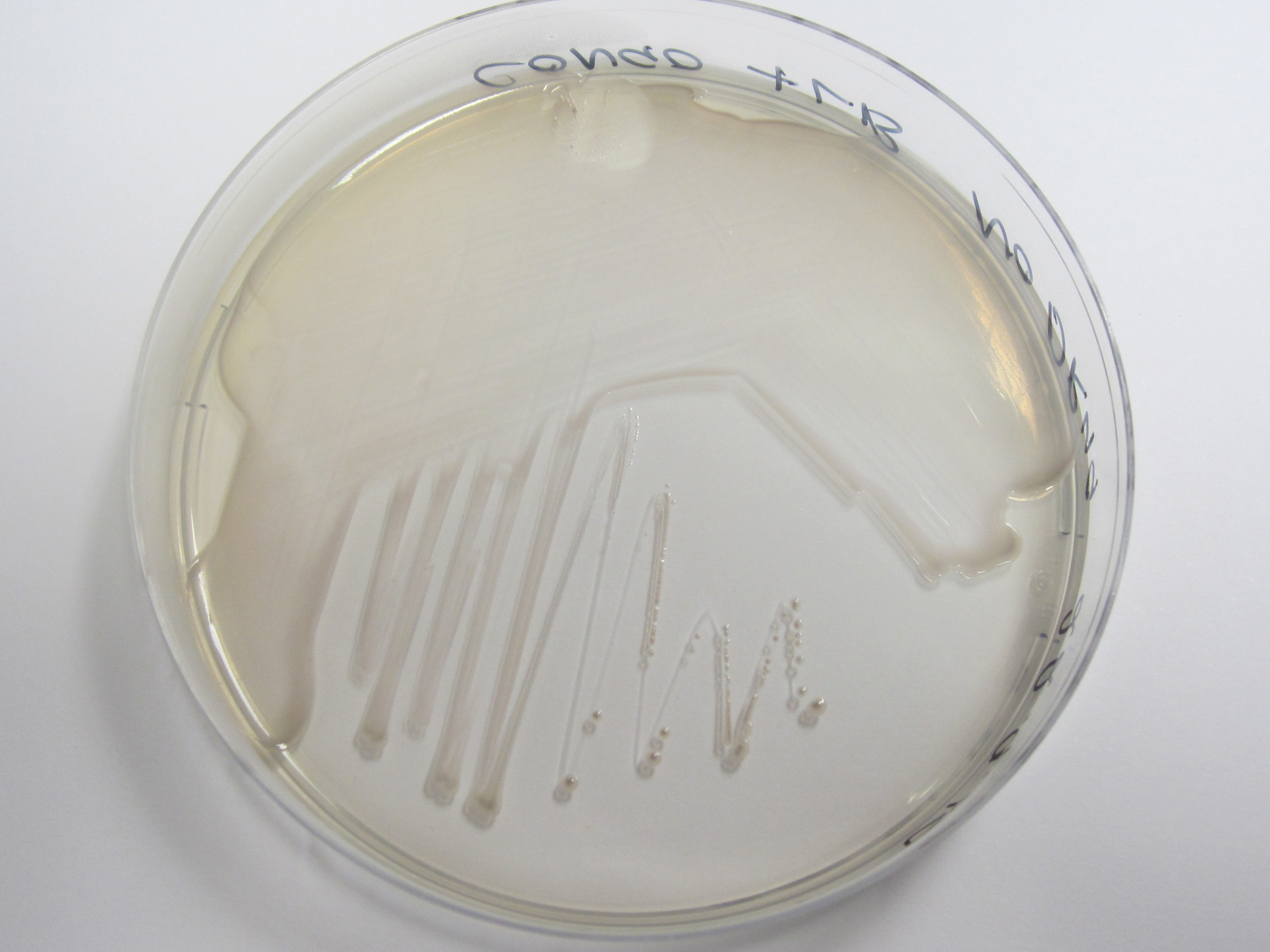 | 
|
From the plates above we can see that our BL21 control that does not have T7+curli insert and does not appear red. However, BL21 that has T7+curli insert appears red. BL21 with a constitutive promoter also shows uptake of dye and shoiws red colonies.
Conclusion:
We conclude successful expression of the curli operon as indicated by the red pigmented colonies. Ability to uptake congo red dye resides in the amyloid structure of the appendages and ionic interactions.

Wednesday (26.09.12)
Aim – We would like to determine if our curli producing BBa_K729018 is capable of producing shear resistant biofilms.
1) Cells are grown overnight in 10ml of LB media at 37˚C, 200rpm.
- a) BBa_K729006
- b) W3110 Control
2) Shake flasks are inoculated with 5 ml of cells into LB media (100ml)
- a) Shake flasks contain pre-sterilised polycarbonate discs
3) Shake flasks are run at 37˚C, 200rpm, for 7 hours.
4) Shake flasks are run at 30˚C, 50rpm, for 14 hours.
5) PC discs are removed from flasks, dried, and stained with crystal violet.
6) Discs are sheared with the small scale shear device, using water as the buffer. Rotaion speed varies from 50rpm to 700rpm.
7) Critical shear radius is measured from diameter of shear resistant area.
Conclusion - Our transformed cells are producing shear resistant biofilm, with double the shear resistance of untransformed biofilms, indicating that our BioBrick is working as intended and is capable of producing shear resistant curliated biofilms.
 "
"