Team:Paris-Saclay/Project/Characterisation
From 2012.igem.org
YohannPetiot (Talk | contribs) |
(→Characterisation) |
||
| (21 intermediate revisions not shown) | |||
| Line 40: | Line 40: | ||
[[Category:Team:Paris-Saclay/Project Gemote|2]] | [[Category:Team:Paris-Saclay/Project Gemote|2]] | ||
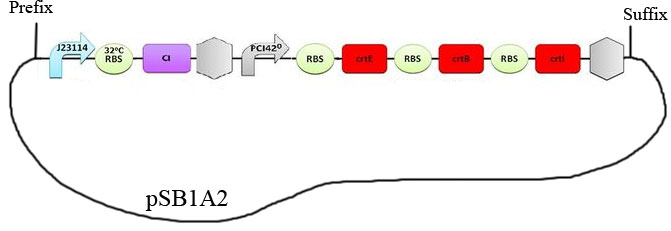
| - | + | Transformed living colonies, that we obtained after Gibson assembly, are supposed to contain the construction detailed below: | |
| - | [[File:Image55.jpeg| | + | [[File:Image55.jpeg|550px|center]] |
The bacteria carrying this plasmid are expected to turn red below 32°C and over 42°C. We used this construction in the characterization experiments. | The bacteria carrying this plasmid are expected to turn red below 32°C and over 42°C. We used this construction in the characterization experiments. | ||
| - | After the Gibson assembly, we obtained many white clones and only one red colony which was named #11. After sequencing, white clones proved to | + | After the Gibson assembly, we obtained many white clones and only one red colony which was named #11. After sequencing, white clones proved to have the backbone plasmid but without the construction. |
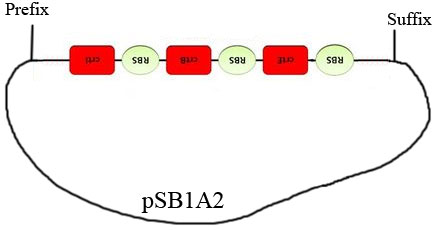
However for clone # 11 , after having received the sequencing results of our construct, we realized that the Gibson Assembly didn’t work as expected. The construct effectively obtained is figured below: | However for clone # 11 , after having received the sequencing results of our construct, we realized that the Gibson Assembly didn’t work as expected. The construct effectively obtained is figured below: | ||
| - | [[File:Image56.jpeg]] | + | [[File:Image56.jpeg|450px|center]] |
We can see that only the part containing the lycopene biosynthesis chain was inserted. Even worse, it was oriented the wrong way around! We had the results from the sequencing recently, so the reason why the part could have been inserted this way remains a mystery that we are still attempting to unravel. The reason why the colonies are red-stained is another strange thing, because there is no promoter before the insertion. The fact that the RBS are still present can however support the argument that a leaky promoting activity could be located upstream from the operon, creating an mRNA. | We can see that only the part containing the lycopene biosynthesis chain was inserted. Even worse, it was oriented the wrong way around! We had the results from the sequencing recently, so the reason why the part could have been inserted this way remains a mystery that we are still attempting to unravel. The reason why the colonies are red-stained is another strange thing, because there is no promoter before the insertion. The fact that the RBS are still present can however support the argument that a leaky promoting activity could be located upstream from the operon, creating an mRNA. | ||
| Line 55: | Line 55: | ||
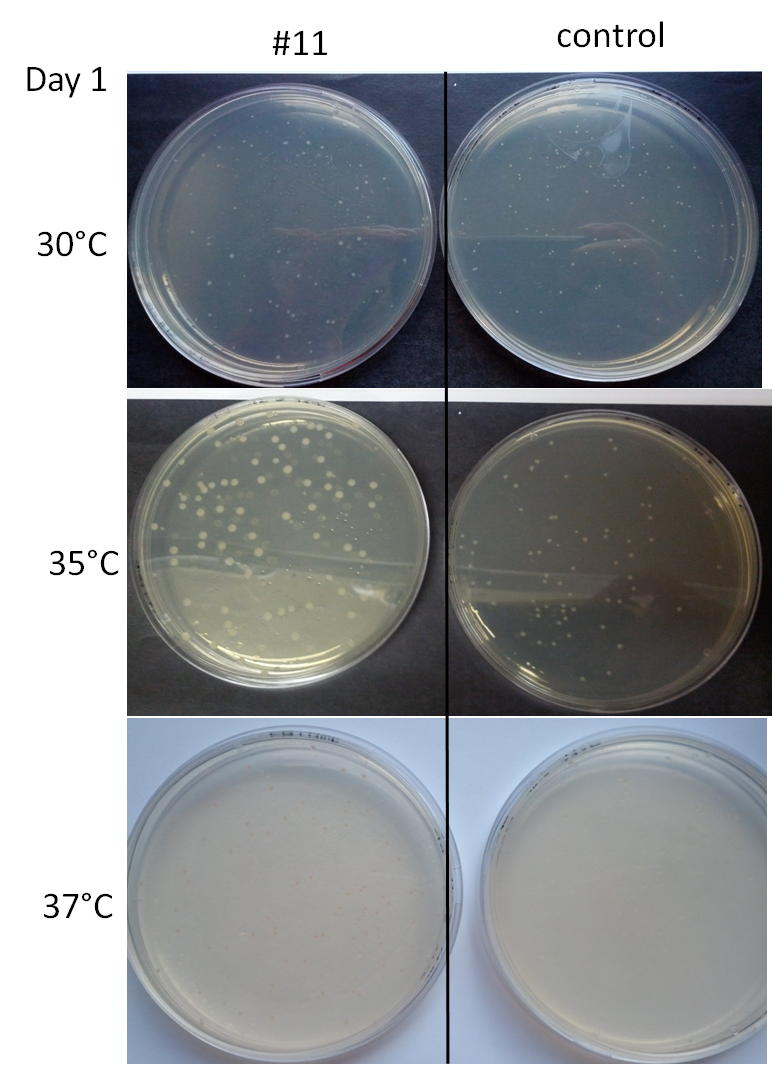
We tested our organism's temperature range for 2 days. The first day, 24 hours after spreading liquid cultures on Petri dishes LB+agar+Ampicilline, we had the following results: | We tested our organism's temperature range for 2 days. The first day, 24 hours after spreading liquid cultures on Petri dishes LB+agar+Ampicilline, we had the following results: | ||
| - | [[File:Image57.png]] | + | [[File:Image57.png|right|380px]] |
| - | [[File:Image58.png]] | + | |
| + | |||
| + | |||
| + | *At 25°C: very small colonies whose color in #11 and in the control was indistinguishable. | ||
| + | *At 28°C: small white colonies for #11 and the control | ||
| + | *At 30°C: white colonies for #11 and the control | ||
| + | *At 35°C: slightly pinkish white colonies for #11, a bit larger than the colonies from the control | ||
| + | *At 37°C: similar colonies for #11 and the control except that the colonies are pink for #11 | ||
| + | *At 40°C: similar to the colonies placed at 37°C for both #11 and the control | ||
| + | *At 42°C: very small colonies for #11 and the control | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
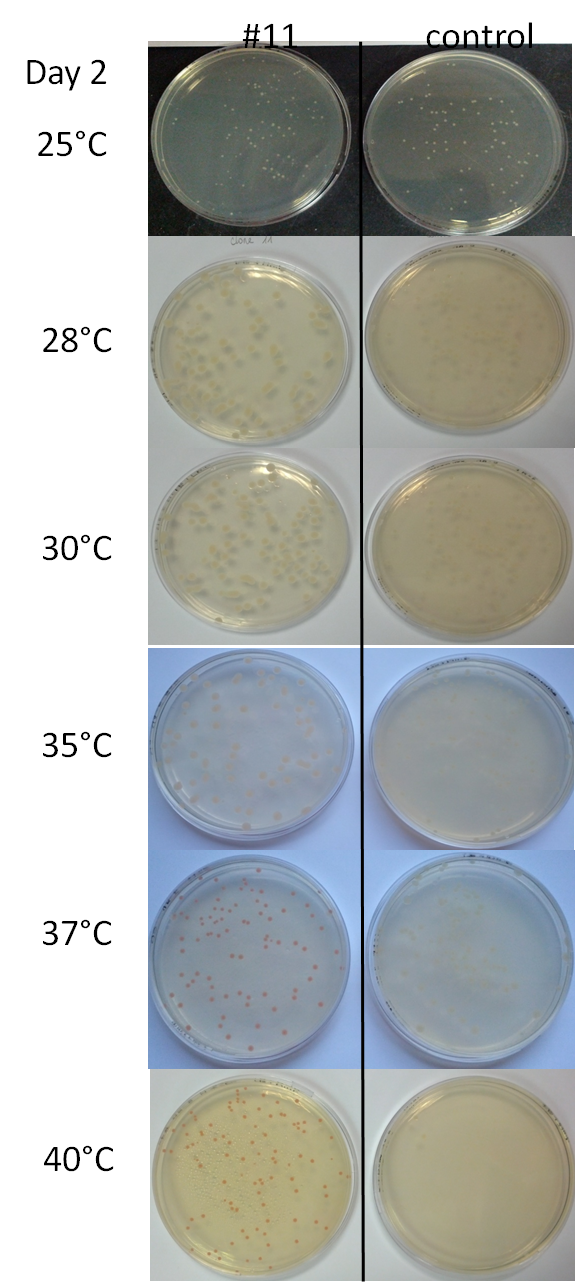
| + | The second day, 48 hours after spreading liquid cultures on Petri dishes, we had the following results: | ||
| + | |||
| + | [[File:Image58.png|right|350px]] | ||
| + | |||
| + | |||
| + | |||
| + | *At 25°C: white colonies for #11 and the control | ||
| + | *At 28°C: white-beige colonies for #11 which are bigger than those of the control | ||
| + | *At 30°C: white-pinkish colonies for #11 which are bigger than those of the control | ||
| + | *At 35°C: pinkish colonies for #11 which are bigger than those of the control | ||
| + | *At 37°C: red colonies for #11 but similar in size than those of the control | ||
| + | *At 40°C: red colonies for #11, the same size than those at 37°C | ||
| + | *At 42°C: for #11, there are different size of colonies, the smallest are red and the biggest are white | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | These experiments allowed us to determine the range of temperature in which colonies turn red. We noticed a faint color at 28°C in 2 days and a gradual pigmentation from light pink to dark pink between 30 and 42 °C. However, we noticed that colonies placed at 42°C are much smaller than those placed at a different temperature. This suggests that at high temperature, lycopene seems to be produced in higher quantity but could disrupt the normal growth of bacteria. This could explain the fact that the colonies don’t develop, even after an incubation of 2 days. | ||
| + | According to the sequencing results, #11 clone contains the lycopene’s biosynthesis pathway biobrick, but no other fragments of RNA thermometer and repressor CI. It is thus surprising to observe a thermo-sensitive production of lycopene. This could be explained by the temperature, which could play a role on the kinetics of the synthesis of the lycopene, through three enzymes of the biosynthesis pathway. | ||
| + | |||
{{Team:Paris-Saclay/Follow}} | {{Team:Paris-Saclay/Follow}} | ||
</div> | </div> | ||
Latest revision as of 01:32, 27 September 2012
Characterisation
Transformed living colonies, that we obtained after Gibson assembly, are supposed to contain the construction detailed below:
The bacteria carrying this plasmid are expected to turn red below 32°C and over 42°C. We used this construction in the characterization experiments. After the Gibson assembly, we obtained many white clones and only one red colony which was named #11. After sequencing, white clones proved to have the backbone plasmid but without the construction. However for clone # 11 , after having received the sequencing results of our construct, we realized that the Gibson Assembly didn’t work as expected. The construct effectively obtained is figured below:
We can see that only the part containing the lycopene biosynthesis chain was inserted. Even worse, it was oriented the wrong way around! We had the results from the sequencing recently, so the reason why the part could have been inserted this way remains a mystery that we are still attempting to unravel. The reason why the colonies are red-stained is another strange thing, because there is no promoter before the insertion. The fact that the RBS are still present can however support the argument that a leaky promoting activity could be located upstream from the operon, creating an mRNA.
In parallel to the sequencing, a test had been done to determine the range of temperature in which clones turned red. Liquid cultures of bacteria were spread on medium containing Ampicillin, then incubated at different temperatures: 25,28,30,35,37,40,42 °C. We observed the colonies color after letting them grow for 2 days. As a control, we used a strain of bacteria containing a plasmid with the Ampicillin resistance in order to control the growth and the color. We tested our organism's temperature range for 2 days. The first day, 24 hours after spreading liquid cultures on Petri dishes LB+agar+Ampicilline, we had the following results:
- At 25°C: very small colonies whose color in #11 and in the control was indistinguishable.
- At 28°C: small white colonies for #11 and the control
- At 30°C: white colonies for #11 and the control
- At 35°C: slightly pinkish white colonies for #11, a bit larger than the colonies from the control
- At 37°C: similar colonies for #11 and the control except that the colonies are pink for #11
- At 40°C: similar to the colonies placed at 37°C for both #11 and the control
- At 42°C: very small colonies for #11 and the control
The second day, 48 hours after spreading liquid cultures on Petri dishes, we had the following results:
- At 25°C: white colonies for #11 and the control
- At 28°C: white-beige colonies for #11 which are bigger than those of the control
- At 30°C: white-pinkish colonies for #11 which are bigger than those of the control
- At 35°C: pinkish colonies for #11 which are bigger than those of the control
- At 37°C: red colonies for #11 but similar in size than those of the control
- At 40°C: red colonies for #11, the same size than those at 37°C
- At 42°C: for #11, there are different size of colonies, the smallest are red and the biggest are white
These experiments allowed us to determine the range of temperature in which colonies turn red. We noticed a faint color at 28°C in 2 days and a gradual pigmentation from light pink to dark pink between 30 and 42 °C. However, we noticed that colonies placed at 42°C are much smaller than those placed at a different temperature. This suggests that at high temperature, lycopene seems to be produced in higher quantity but could disrupt the normal growth of bacteria. This could explain the fact that the colonies don’t develop, even after an incubation of 2 days.
According to the sequencing results, #11 clone contains the lycopene’s biosynthesis pathway biobrick, but no other fragments of RNA thermometer and repressor CI. It is thus surprising to observe a thermo-sensitive production of lycopene. This could be explained by the temperature, which could play a role on the kinetics of the synthesis of the lycopene, through three enzymes of the biosynthesis pathway.
 "
"



Follow us !