Team:University College London/LabBook/Week9
From 2012.igem.org
Sednanalien (Talk | contribs) (→Friday 10.8.12) |
Rwilkinson (Talk | contribs) (→Friday 10.8.12) |
||
| (41 intermediate revisions not shown) | |||
| Line 330: | Line 330: | ||
! Antibiotic Stock !! Presumed Concentration | ! Antibiotic Stock !! Presumed Concentration | ||
|- | |- | ||
| - | | Kanamycin | + | | Kanamycin 01 || 50ug/ml |
|- | |- | ||
| - | | | + | | Chloramphenicol 01 || 50ug/ml |
|- | |- | ||
| - | | Tetracycline | + | | Chloramphenicol 02 || 50ug/ml |
| + | |- | ||
| + | | Chloramphenicol 03 || 50ug/ml | ||
| + | |- | ||
| + | | Tetracycline 01 || 50ug/ml | ||
| + | |- | ||
| + | | Tetracycline 02|| 50ug/ml | ||
| + | |- | ||
| + | | Tetracycline 03 || 50ug/ml | ||
|- | |- | ||
| Ampicillin 01 || 50ug/ml | | Ampicillin 01 || 50ug/ml | ||
| Line 354: | Line 362: | ||
| - | + | The following table summarises the type of plates and their contents. Top10, W3110 and pTopDsh have no antibiotic resistance, and so they are our negative controls – there should be no growth except on drug-free agar. WNu has Ampicillin resistant, and should go in the presence of Ampicillin but not other antiobiotics. pTopDsh carries Tetracycline resistance, and should grow in the presence of Tetracycline but not the other antibiotics. | |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| - | ! Antibiotic Stock !! Sample Added !! Expected Result | + | ! Antibiotic Stock !! Sample Added !! Expected Result !! Result |
| + | |- | ||
| + | |rowspan="2"| Kanamycin 01 || WNu || No growth (WNu is Ampicillin resistant only) || no colony | ||
| + | |- | ||
| + | | WNu|| No growth (WNu is Ampicillin resistant only)|| no colony | ||
| + | |- | ||
| + | |rowspan="2"| Chloramphenicol 01 || Cobalt Curli Cells || Growth (Cobalt Curli cells is Chloramphenicol resistant) || colonies | ||
| + | |- | ||
| + | | WNu || No growth (WNu is Ampicillin resistant only)|| no colony | ||
| + | |- | ||
| + | |rowspan="2"| Chloramphenicol 02 ||Cobalt Curli Cells || Growth (Cobalt Curli cells is Chloramphenicol resistant) || colonies | ||
| + | |- | ||
| + | | WNu || No growth (WNu is Ampicillin resistant only)|| no colony | ||
| + | |- | ||
| + | |rowspan="2"| Chloramphenicol 03 || Cobalt Curli Cells|| Growth (Cobalt Curli cells is Chloramphenicol resistant) || colonies | ||
| + | |- | ||
| + | |W3110 || No growth (W3110 has no resistance)|| no colony | ||
| + | |- | ||
| + | |rowspan="2"| Tetracycline 01|| WNu ||No growth (WNu is Ampicillin resistant only)|| no colony | ||
| + | |- | ||
| + | |75 ||Growth (75 is Tetracycline Resistant)|| colonies | ||
|- | |- | ||
| - | | rowspan="2"| | + | |rowspan="2"| Tetracycline 02|| WNu ||No growth (WNu is Ampicillin resistant only)|| no colony |
| - | + | ||
| - | |WNu ||No growth (WNu is Ampicillin resistant only) | + | |
|- | |- | ||
| - | + | |75 ||Growth (75 is Tetracycline Resistant)|| colonies | |
| - | + | ||
| - | |75 || Growth (75 is Tetracycline Resistant) | + | |
|- | |- | ||
| - | |rowspan="2"| Tetracycline | + | |rowspan="2"| Tetracycline 03|| WNu ||No growth (WNu is Ampicillin resistant only)|| no colony |
|- | |- | ||
| - | |75 ||Growth (75 is Tetracycline Resistant) | + | |75 ||Growth (75 is Tetracycline Resistant)|| no colony |
|- | |- | ||
| - | |rowspan="2"| Ampicillin 01 || pTopDsh || No Growth (pTopDsh has no resistance) | + | |rowspan="2"| Ampicillin 01 || pTopDsh || No Growth (pTopDsh has no resistance)|| no colony |
|- | |- | ||
| - | |WNu||No growth (WNu is Ampicillin resistant only) | + | |WNu||No growth (WNu is Ampicillin resistant only)|| colonies |
|- | |- | ||
| - | | rowspan="2"| Ampicillin 02|| WNu || Growth (WNu is Ampicillin resistant) | + | | rowspan="2"| Ampicillin 02|| WNu || Growth (WNu is Ampicillin resistant)|| no colony |
|- | |- | ||
| - | |W3100 || No Growth (W3100 has no resistance) | + | |W3100 || No Growth (W3100 has no resistance)|| colonies |
|- | |- | ||
| - | | rowspan="2"| Ampicillin 03 || WNu ||Growth (WNu is Ampicillin resistant) | + | | rowspan="2"| Ampicillin 03 || WNu ||Growth (WNu is Ampicillin resistant)|| no colony |
|- | |- | ||
| - | | pTopDsh || No Growth (pTopDsh has no resistance) | + | | pTopDsh || No Growth (pTopDsh has no resistance)|| no colony |
|- | |- | ||
| - | |rowspan="2"| Ampicillin 04 || WNu ||Growth (WNu is Ampicillin resistant) | + | |rowspan="2"| Ampicillin 04 || WNu ||Growth (WNu is Ampicillin resistant)|| colonies |
|- | |- | ||
| - | | pTopDsh || No Growth (pTopDsh has no resistance) | + | | pTopDsh || No Growth (pTopDsh has no resistance)|| no colony |
|- | |- | ||
| - | |rowspan="5"| Control - No Drug || W3110 || Growth | + | |rowspan="5"| Control - No Drug || W3110 || Growth||colonies |
|- | |- | ||
| - | | Top10 || Growth | + | | Top10 || Growth|| colonies |
|- | |- | ||
| - | | 75 || Growth | + | | 75 || Growth|| colonies |
|- | |- | ||
| - | | CoCurli || Growth | + | | CoCurli || Growth|| colonies |
|- | |- | ||
| - | |pTopDsh || Growth | + | |pTopDsh || Growth|| colonies |
|} | |} | ||
| - | + | Plates results are as expected, no problem with the activity of the antibiotics however we are not sure if the antibiotic are at the concentration stated. | |
<html> | <html> | ||
Latest revision as of 03:51, 27 September 2012



Contents |
Monday 6.7.12
Aim: Inoculate the Plates that were transformed on Thursday. On Thursday we began Expt 8.5 by transforming the BioBricks, and our results on Friday indicated there was growth for all. (However, the negative control was contaminated, so we will have to be careful to assess the analytical restriction digest for correct products). The table below describes the BioBricks that were used:
The two constitutive promoters were used because we were never able to detect our previous constitutive promoter BBa_J23119 (Expt 7.3) and our 3A ligation of BBa_J23119 and BBa_B0034 failed (Expt 8.4). This does not necessarily indicate our transformation of BBa_J23119 has failed, but we decided to transform several more in case. The RBS promoter BBa_B0030 was transformed for the same reason. The Gas Vesicle Cluster was transformed because all previous attempts have failed.
Method
Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Step 2 – Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic required for each BioBrick.
| Samples | Volume Inoculated | Broth (ml) | Antibiotic (ug/ml) | |
|---|---|---|---|---|
| BioBrick | BBa_J23100 | 10ul | Lysogeny Broth (5) | Ampicillin(50ug/ml) |
| 90ul | ||||
| BBa_J23106 | 10ul | |||
| 90ul | ||||
| BBa_B0030 | 10ul | |||
| 90ul | ||||
| BBa_I750016 | 10ul | |||
| 90ul | ||||
Tuesday 7
Aim: Results from Colony Picking
ResultsThe table below indicates there was growth for all BioBricks.
| Biobrick | Growth |
|---|---|
| I750016 sample 1 | No Growth |
| I750016 sample 2 | No Growth |
| I750016 sample 3 | No Growth |
| B0030 sample 1 | No Growth |
| B0030 sample 2 | No Growth |
| B0030 sample 3 | No Growth |
| J23100 sample 1 | No Growth |
| J23100 sample 2 | No Growth |
| J23100 sample 3 | No Growth |
| J23106 sample 1 | Growth |
| J23106 sample 1 | Growth |
| J23106 sample 1 | Growth |
Conclusion: We are unsure why we have failed to get colonies for many of the BioBricks. Again, we are considering the possibility that our agar is not selective enough. Initially we thought this may be due to adding Antibiotic while the agar is too hot, leading to degradation of the sample. However this has been corrected in the protocol, but the problem is persisting. Instead we are considering running a troubleshoot by testing the selectivity of the various eppendorfs of antibiotic stocks we made at the beginning of the summer. If any show less selectivity then we can stop using them. This will be considered for later in the week. Meanwhile, we can miniprep and nanodrop BBa_J23106 (strong constitutive promoter)
Step 1 - Pellet Cells: Pellet 1.5-5ml bacterial culture containing the plasmid by centrifugation g = 6000
Time = 2 min
Temperature = (15-25oC)
Step 2 - Resuspend Cells: Add 250ul S1 to the pellet and resuspend the cells completely by vortexing or pipetting. Transfer the suspension to a clean 1.5ml microcentrifuge tube.
Step 3 - Puncturing Cell Membrane: Add 250ul S2 gently mix by inverting the tube 4-6 times to obtain a clear lysate. Incubate on ice or at room temperature for NOT longer than 5 min.
Step 4 - Neutralising S2: Add 400ul Buffer NB and gently mix by inverting the tube 6-10 times, until a white precipitate forms.
Step 5 - Centrifuge:
g: 14000
Time:10 minutes
Temperature: (15-25oC)
Step 6 - Centrifuge: Transfer the supernatant into a column assembled in a clean collection tube (provided. Centrifuge:
g = 10,000
Time: 1 minute
Temperature: (15-25oC)
Step 7 - Wash Column: Discard the flow-through and wash the spin column by adding 700ul of Wash Buffer. Centrifuge:
g - 10,000
Time - 1 minute
Temperature: (15-25oC)
Step 8 - Remove residual ethanol: Centrifuge:
g - 10,000
Time - 1 minute
Temperature: (15-25oC)
Step 9 - Elute DNA: Place the column into a clean microcentrifuge tube. Add 50-100ul of Elution Buffer, TE buffer or sterile water directly onto column membrane and stand for 1 min. Centrifuge:
g - 10,000
Time - 1 minute
Temperature: (15-25oC)
Step 10 - Storage: Store DNA at 4oC or -20oC
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: There was no product for BBa_J23106 on the gel, therefore either the transformation failed or the concentration is very low

Monday 6.7.12
Aim: Repeat the inoculation of the ligation products (J23119 + B0034) done last week. Innoculation is only done for the (J23119 + B0034), but not for the (starvation promoter + B0034) because that ligation did not transform.
Method
Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Step 2 – Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic required for each BioBrick.
| Ligation | No. innoculations | Broth | Antibiotic |
|---|---|---|---|
| J23119 + B0034 | 3 | Lysogeny Broth | Kanamycin |
Tuesday 7
Aim: Check results of Colony Picking
Results: The table below indicates whether there was growth for the BioBricks ligations
| Ligation | Growth/No Growth |
|---|---|
| J23119+B0034 sample 1 | Growth |
| J23119+B0034 sample 2 | Growth |
| J23119+B0034 sample 3 | Growth |
Conclusion: We can proceed now to miniprep the samples, and run them on a gel to test the presence of a band
Method:
Step 1 - Pellet Cells: Pellet 1.5-5ml bacterial culture containing the plasmid by centrifugation g = 6000
Time = 2 min
Temperature = (15-25oC)
Step 2 - Resuspend Cells: Add 250ul S1 to the pellet and resuspend the cells completely by vortexing or pipetting. Transfer the suspension to a clean 1.5ml microcentrifuge tube.
Step 3 - Puncturing Cell Membrane: Add 250ul S2 gently mix by inverting the tube 4-6 times to obtain a clear lysate. Incubate on ice or at room temperature for NOT longer than 5 min.
Step 4 - Neutralising S2: Add 400ul Buffer NB and gently mix by inverting the tube 6-10 times, until a white precipitate forms.
Step 5 - Centrifuge:
g: 14000
Time:10 minutes
Temperature: (15-25oC)
Step 6 - Centrifuge: Transfer the supernatant into a column assembled in a clean collection tube (provided. Centrifuge:
g = 10,000
Time: 1 minute
Temperature: (15-25oC)
Step 7 - Wash Column: Discard the flow-through and wash the spin column by adding 700ul of Wash Buffer. Centrifuge:
g - 10,000
Time - 1 minute
Temperature: (15-25oC)
Step 8 - Remove residual ethanol: Centrifuge:
g - 10,000
Time - 1 minute
Temperature: (15-25oC)
Step 9 - Elute DNA: Place the column into a clean microcentrifuge tube. Add 50-100ul of Elution Buffer, TE buffer or sterile water directly onto column membrane and stand for 1 min. Centrifuge:
g - 10,000
Time - 1 minute
Temperature: (15-25oC)
Step 10 - Storage: Store DNA at 4oC or -20oC
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: There was no product on the gel, suggesting the ligation failed.

Monday 8.8.12
Aim: To repeat Expt 8.2, where we undertook PCR amplification of pSB1A3, pSB1C3, pSB1K3 for 3A assembly. While we achieved bands of the correct size for all three plasmid backbones in Expt 8.2, the nanodrop concentration for pSB1A3 and pSB1C3 was very low (unusable). Therefore we decided to repeat the PCR. As the concentration of pSB1K3 was not particularly high, we decided to repeat this again also.
Method:
Step 1 - Setting up PCR tubes: Thaw reagents and add to PCR tubes in the proportions described in the table below
| PCR Components | Volume (ul) |
|---|---|
| 5x Reaction Buffer | 10 |
| 25mM MgCl2 | 4 |
| 10mM dNTPs | 1 |
| 10uM Forward primer | 5 |
| 10uM Reverse primer | 5 |
| DNA Polymerase | 0.25 |
| Nuclease Free Water | 24.25 |
| Template DNA | 0.5 |
| Total Volume | 0.5 |
Step 2 - PCR program: Add PCR tubes to a thermocycler and run under the following conditions.
| PCR conditions | Temp (oC) | Time (s) |
|---|---|---|
| Initial Denaturation (1 cycle) | 95 | 30 |
| Denaturation/Annealing/Extension (30 cycles) | 95/55/72 | 10/25/120 |
| Final Extension (1 cycle) | 72 | 600 |
| Hold | 4 | ∞ |
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: The image below shows a 1% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 9.1, with a 1000bp ladder. No products were detected. We expected a product corresponding to the size of the plasmid pSB1K3 (2000bp), pSB1C3 (2000bp) and pSB1A3 (2000bp) in Lanes 1-3 respectively. These are indicated by the letters A (pSB1K3), B (pSB1C3) and C (pSB1A3).

| Backbone | Expected Size |
|---|---|
| pSB1K3 | 2204bp |
| pSB1C3 | 2070bp |
| pSB1A3 | 2155bp |
Conclusion: We feel the poor nanodrop results from Expt 8.2, and the poor electrophoresis results from this Expt may be a result of the choice of polymerase. Our supervisors recommended we try Phusion Polymerase, which we will attempt tomorrow.
Tuesday 9.8.12
Aim: To repeat PCR with Phusion Polymerase to amplify the three plasmid backbones. As we had more confidence that this PCR reaction would work, we included an extra plasmid, pSB1T3, also required for ligation.
Method
Step 1 - Setting up PCR tubes: Thaw reagents and add to PCR tubes in the proportions described in the table below
| PCR Components | Volume (ul) |
|---|---|
| 5x Reaction Buffer | 10 |
| 25mM MgCl2 | 4 |
| 10mM dNTPs | 1 |
| 10uM Forward primer | 5 |
| 10uM Reverse primer | 5 |
| DNA Polymerase | 0.25 |
| Nuclease Free Water | 24.25 |
| Template DNA | 0.5 |
| Total Volume | 0.5 |
Step 2 - PCR program: Add PCR tubes to a thermocycler and run under the following conditions.
| PCR conditions | Temp (oC) | Time (s) |
|---|---|---|
| Initial Denaturation (1 cycle) | 95 | 30 |
| Denaturation/Annealing/Extension (30 cycles) | 95/55/72 | 10/25/120 |
| Final Extension (1 cycle) | 72 | 600 |
| Hold | 4 | ∞ |
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: The image below shows a 1% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 9.1, with a 1000bp ladder. In Lane 1 can be seen a product corresponding to the correct size of pSB1A3 (2155) as indicated by A. In Lane 2 is a product, shown by B, which is of the correct size for the plasmid pSB1K3 (22040bp). In Lane 3, we expect a band of size 2461bp, for the plasmid pSB1T3. This is shown by C. As it is absent, it would appear the PCR reaction failed. In Lane 4 is a product corresponding to pSB1C3 (2070bp) as shown by D.

| Backbone | Expected Size |
|---|---|
| pSB1K3 | 2204bp |
| pSB1C3 | 2070bp |
| pSB1A3 | 2155bp |
| pSB1T3 | 2461bp |
Conclusion: The PCR reaction worked for pSB1K3, pSB1C3 and pSB1A3. This would suggest the Phusion Polymerase is a better choice than Taq polymerase. These plasmids can now be nanodropped, and if their concentration is sufficient they can be used for ligation.
Thursday 10.8.12
Aim: To carry out nanodrop for the PCR reaction of plasmids with phusion polymerase
Method
Software ND-1000 Model:
Step 1: Initialise the spectrophotometer by pipetting 1 µ of clean water onto lower optic surface, lowering the lever arm and selecting ‘initialise’ in the ND-1000 software
Step 2: Wipe and add elution buffer as negative control. Click blank in ND-1000 software
Step 3: Wipe and add 1 µl sample
Step 4: On the software set lambda to 260nm
Step 5: Lower the lever arm and click measure in ND-1000 software
Step 6: Take readings for concentration and purity
Step 7: Once measurement complete, wipe surface
| Plasmid | λ260 | λ 280 |
|---|---|---|
| pSB1A3 (ng/μl) | 45.7 | 45.4 |
| pSB1C3 (ng/μl) | 52.2 | 48.0 |
| pSB1K3 (ng/μl) | 57.9 | 59.4 |
Conclusion: The results of the nanodrop indicate that we have usable concentrations of the plasmids, though they are not particularly high. However, we can attempt to use these for 3A ligation.

Wednesday 8.8.12
Aim: To extract the Laccase gene for the Degradation module and the IrrE gene for the Salt Tolerance module.
Method:
DNA for the laccase gene was extracted from cultured colonies according to the protocol below:
Step 1 - Picking Colony: Using a pipette select an isolated colony
Step 2 - Deposit: Dip the colony in 10ul of dH20, preferably in screw top container
Step 3 - Boiling Stage:Boil the dH20 100 for 5-10mins
Step 4 - Centrifuge:
Time: 1 min
RPM: 8000
Temp: 18oC
Step 5 - Remove Supernatant: The supernatant contains the DNA template.
DNA for IrrE was extracted by Genomic Extraction
Protocol: Genomic Extraction
Primers should be designed to:
1) Be 18-25 nucleotides in length
2) Have a melting temperature that does not differ by more than 5oC between the two primers of a pair
3) GC nucleotides should constitute 40-60%
4) Have a 3’primer end that is not complimentary to any region of the primer pair
5) Contain an equal distribution of the nucleotides
6) Closely match the target sequence (and so known polymorphic sites are avoided)
7) Have a GC clamp on either end
The design of the primers is shown by the table below
Step 1 - Setting up PCR tubes: Thaw reagents and add to PCR tubes in the proportions described in the table below
| PCR Components | Volume (ul) |
|---|---|
| 5x Reaction Buffer | 10 |
| 25mM MgCl2 | 4 |
| 10mM dNTPs | 1 |
| 10uM Forward primer | 5 |
| 10uM Reverse primer | 5 |
| DNA Polymerase | 0.25 |
| Nuclease Free Water | 24.25 |
| Template DNA | 0.5 |
| Total Volume | 0.5 |
Step 2 - PCR program: Add PCR tubes to a thermocycler and run under the following conditions.
| PCR conditions | Temp (oC) | Time (s) |
|---|---|---|
| Initial Denaturation (1 cycle) | 95 | 30 |
| Denaturation/Annealing/Extension (30 cycles) | 95/55/72 | 10/25/120 |
| Final Extension (1 cycle) | 72 | 600 |
| Hold | 4 | ∞ |
Step 1 - Setting up PCR tubes The table below gives the identity of the primers used for each reaction.
| DNA Extracted | Function | Module | Primer Pair | Primer | Primer Sequence |
|---|---|---|---|---|---|
| BBa_K729002 | Laccase Gene | Degradation | LR1/LF1 | LR1 | gaatacggtctttttataccg |
| LF1 | gaaataactatgcaacgtcg | ||||
| REVLF2/LFTW0 | REVLF2 | gtttcttcctgcagcggccgctactagtagaatacggtctttttataccg | |||
| LFTW0 | gtttcttcgaattcgcggccgcttctagaggaaataactatgcaacgtcg | ||||
| BBa_K729001 | IrrE | Salt Tolerance | STF1/ST2R | STF1 | atggggccaaaagctaaagctgaagcc |
| ST2R | tcactgtgcagcgtcctgcg | ||||
| STF3/ST4R | STF3 | gtttcttcgaattcgcggccgcttctagagatggggccaaaagctaaagctgaagcc | |||
| ST4R | gtttcttcctgcagcggccgctactagtatcactgtgcagcgtcctgcg |
Thursday 9.8.12
Aim: Run a gel for the PCR reaction of irrE and Laccase carried out yesterday.
Method:
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: The image belows shows that the gel failed to produce any products of the correct size. We feel the best explanation of this is that there was human error somewhere along the lines – leading to a mix up of the samples.

Thursday 9.8.12
Aim: inoculation of marine bacterium. We aim to grow up a stock of the marine bacterium Oceanibulbus indolifex, which has been selected as one of the chassis we wish to investigate.
Method
Oceanibulbus was ordered from NCMB and recieved as a liquid stock.
Step 1: Prepare agar plates
Step 2: Dip an inoculation loop into the liquid stock of bacteria and spread over the surface of the agar
Step 3: Leave the bacteria to grow overnight or over the weekend.
Friday 10.8.12
Aim: Check the results of O.indolifex culture
Results: The image below indicates there was growth for both inoculations of O.indolifex.
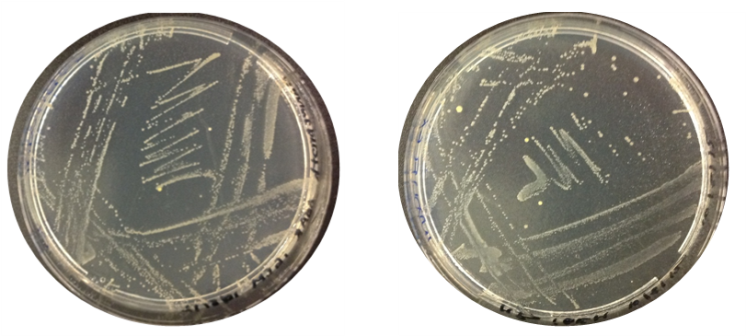

Thursday 9.8.12
Aim: To check the selectivity of our antibiotic stocks. After growth on agar plates, our transformations frequently fail to grow after inoculation into falcons. This has led us to consider the possibility that the selection has not been strong enough, leading to the growth of untransformed cells. We have designed an experiment to test whether our stocks of antibiotics are effective enough. Numerous agar plates will be made with varying antibiotic resistance, and will be inoculated with a variety of antibiotic-resistant and non-resistant bacteria. If non-resistant bacteria can grow on the antibiotic-inoculated agar, we will have some indication of whether the antibiotic activity is high enough.
Method:
Making plates
The table below indicates the plates that were made with each antibiotic. NB// we have several eppendorf stocks of antibiotics; the plate was labelled according to the eppendorf stock it came from. This is intended to identify any of our stocks which may not be of sufficient quality to create a selective agar.
| Antibiotic Stock | Presumed Concentration |
|---|---|
| Kanamycin 01 | 50ug/ml |
| Chloramphenicol 01 | 50ug/ml |
| Chloramphenicol 02 | 50ug/ml |
| Chloramphenicol 03 | 50ug/ml |
| Tetracycline 01 | 50ug/ml |
| Tetracycline 02 | 50ug/ml |
| Tetracycline 03 | 50ug/ml |
| Ampicillin 01 | 50ug/ml |
| Ampicillin 02 | 50ug/ml |
| Ampicillin 03 | 50ug/ml |
| Ampicillin 04 | 50ug/ml |
Friday 10.8.12
Aim: To set up our samples on the plates made yesterday
Method: the plates from yesterday have been split into half (in order to prevent wasting resources). Each side is being treated as a separate sample.
The following table summarises the type of plates and their contents. Top10, W3110 and pTopDsh have no antibiotic resistance, and so they are our negative controls – there should be no growth except on drug-free agar. WNu has Ampicillin resistant, and should go in the presence of Ampicillin but not other antiobiotics. pTopDsh carries Tetracycline resistance, and should grow in the presence of Tetracycline but not the other antibiotics.
| Antibiotic Stock | Sample Added | Expected Result | Result |
|---|---|---|---|
| Kanamycin 01 | WNu | No growth (WNu is Ampicillin resistant only) | no colony |
| WNu | No growth (WNu is Ampicillin resistant only) | no colony | |
| Chloramphenicol 01 | Cobalt Curli Cells | Growth (Cobalt Curli cells is Chloramphenicol resistant) | colonies |
| WNu | No growth (WNu is Ampicillin resistant only) | no colony | |
| Chloramphenicol 02 | Cobalt Curli Cells | Growth (Cobalt Curli cells is Chloramphenicol resistant) | colonies |
| WNu | No growth (WNu is Ampicillin resistant only) | no colony | |
| Chloramphenicol 03 | Cobalt Curli Cells | Growth (Cobalt Curli cells is Chloramphenicol resistant) | colonies |
| W3110 | No growth (W3110 has no resistance) | no colony | |
| Tetracycline 01 | WNu | No growth (WNu is Ampicillin resistant only) | no colony |
| 75 | Growth (75 is Tetracycline Resistant) | colonies | |
| Tetracycline 02 | WNu | No growth (WNu is Ampicillin resistant only) | no colony |
| 75 | Growth (75 is Tetracycline Resistant) | colonies | |
| Tetracycline 03 | WNu | No growth (WNu is Ampicillin resistant only) | no colony |
| 75 | Growth (75 is Tetracycline Resistant) | no colony | |
| Ampicillin 01 | pTopDsh | No Growth (pTopDsh has no resistance) | no colony |
| WNu | No growth (WNu is Ampicillin resistant only) | colonies | |
| Ampicillin 02 | WNu | Growth (WNu is Ampicillin resistant) | no colony |
| W3100 | No Growth (W3100 has no resistance) | colonies | |
| Ampicillin 03 | WNu | Growth (WNu is Ampicillin resistant) | no colony |
| pTopDsh | No Growth (pTopDsh has no resistance) | no colony | |
| Ampicillin 04 | WNu | Growth (WNu is Ampicillin resistant) | colonies |
| pTopDsh | No Growth (pTopDsh has no resistance) | no colony | |
| Control - No Drug | W3110 | Growth | colonies |
| Top10 | Growth | colonies | |
| 75 | Growth | colonies | |
| CoCurli | Growth | colonies | |
| pTopDsh | Growth | colonies |
Plates results are as expected, no problem with the activity of the antibiotics however we are not sure if the antibiotic are at the concentration stated.

Friday 10.8.12
Aim: To characterise the unmodified Curli BioBrick. As part of our experiment, we want to change the promoter in front of the Curli cluster of genes. This experiment will test the protocol we intend to use using the original BioBrick (BBa_K540000)
Method
Congo Red
Step 1 - Prepare: Set out the Congo Red plates already prepared.
Step 2 - Streak Cells: Streak a sample of control cells (cells not expressing Curli) onto one antiobiotic-free congo red plate, and onto one antibiotic-inoculated congo red plate. Do the same for the curli-expressing cell line.
Step 3 - Incubate: Incubate cells overnight at 37oC or over the weekend at 30oC.
Step 2 - Streak Cells: Streak a sample of WNu onto a Antibiotic-inoculated Congo Red plate and a Antibiotic-Free Congo Red Plate. Do the same for the CoCurli (BBa_K540000 transformed) cells. The table below indicates the results expected for each of these four plates.
| Cell Type | Expected Results on Antibiotic-free Congo Red Agar | Expected Results on Chloramphenicol-inoculated Congo Red Agar |
|---|---|---|
| WNu | Formation of White Colonies | No Colony Formation (WNu are not Chloramphenicol-resistant) |
| K540000 (cobalt curli) transformed cells | Formation of Red Colonies | Formation of Red Colonies |
Step 3 - Incubate: A sample of each were cultured at 30 degrees over the weekend because Curli production is expressed better at lower temperatures.

Friday 10.8.12
Aim: To assay the nuclease activity in a test of the protocol. We will use a cell line called WNu which has native secreted nuclease activity. This, alongside a nuclease-negative control, will be cultured on agar containing DNA. Nuclease activity will be indicated by the presence of 'halos' surrounding the colonies, where nuclease activity has degraded DNA and increased the translucence of the agar.
Method
Prepare DNAase agar +IPTG + amp.
Results
The table below summarises the results from this experiment. Also included is an image of the agars, with the halo of nuclease activity indicated.
| Sample | Ampicillin + IPTG | Ampicillin | No Antibiotic |
|---|---|---|---|
| Control BBa_K540000 transformed cells | Opaque | Opaque | Opaque |
| WNu Cells | Halo Formation | Halo Formation | Halo Formation |

 "
"