|
|
| (46 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | <!-- Include the next line at the beginning of every page --> | | <!-- Include the next line at the beginning of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_eppis.resized.jpg|3}} | | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_eppis.resized.jpg|3}} |
| | + | [[File:LMU Glow Spore2 cutII.jpg|620px|link=]] |
| | | | |
| | | | |
| - | [[File:LMU Glow Spore2 cut.jpg|620px]] | + | [[File:SporeCoat.png|100px|right|link=]] |
| | | | |
| - | [[File:SporeCoat.png|100px|right|link=Team:LMU-Munich/Spore_Coat_Proteins]]
| |
| | | | |
| | | | |
| | + | =='''Sporo'''beads - What Protein Do ''You'' Want to Display?== |
| | + | <br> |
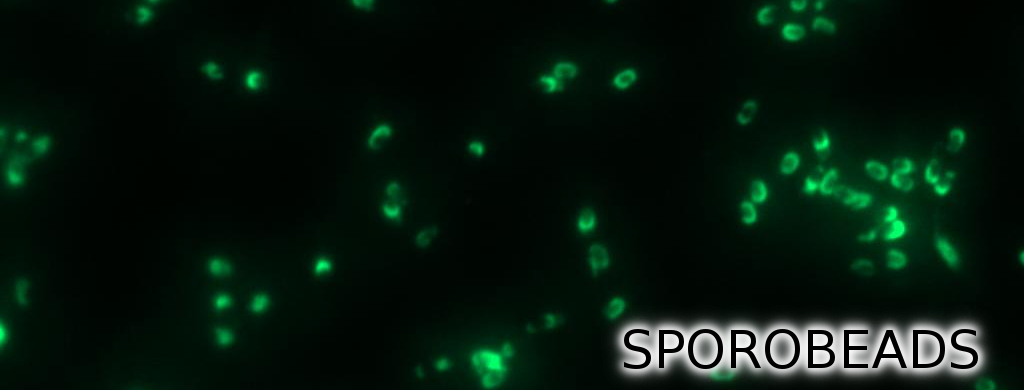
| | + | <p align="justify">'''Sporo'''beads are the first generation of ''Bacillus subtilis'' endospores displaying a protein of our choice on their outermost layer, the spore crust. In future '''Sporo'''beads could serve as a platform for protein display and thus be used for numerous versatile [https://2012.igem.org/Team:LMU-Munich/Application applications]. Our goal was to show that ''B. subtilis'' spores have the ability to do so. As a [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins/result proof of principle] we successfully fused GFP to the spore crust and obtained fluorescence with microscopy.</p> |
| | | | |
| | | | |
| - | =='''Sporo'''beads - what protein do ''you'' want to display?== | + | <div class="box"> |
| - | | + | ===Scientific Background=== |
| - | | + | {| "width=100%" style="text-align:center;" style="align:right"| |
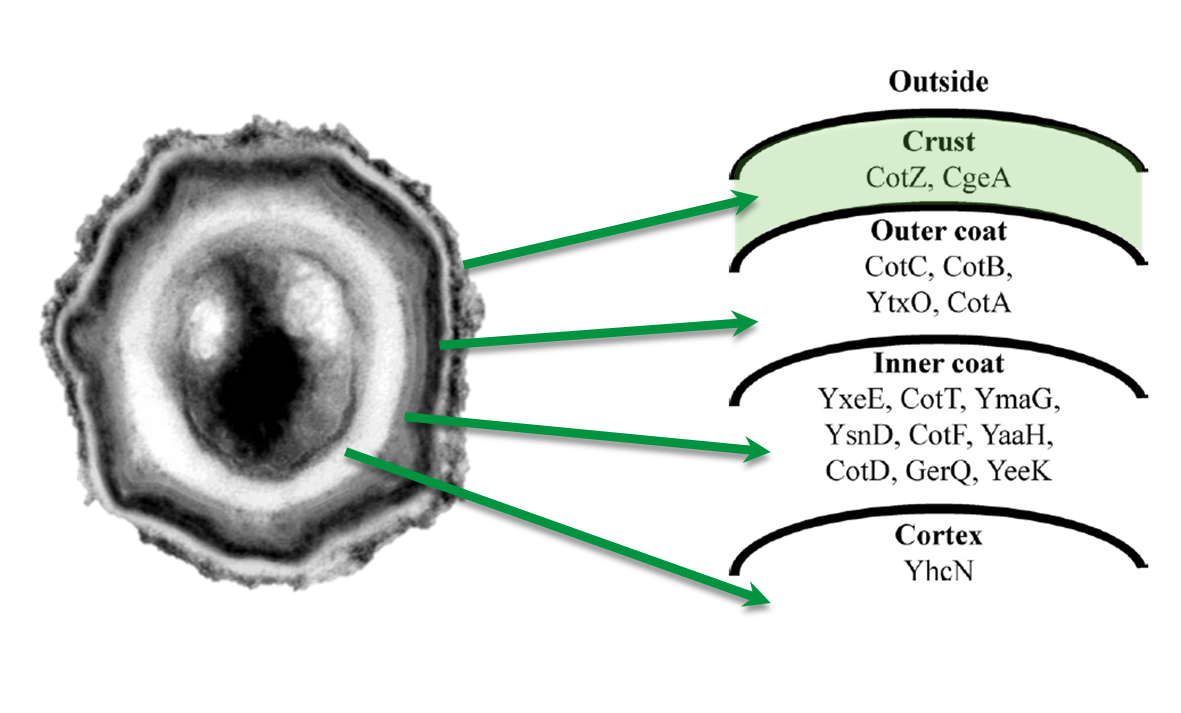
| - | <p align="justify">'''Sporo'''beads are ''Bacillus subtilis'' spores modulated in a way that they can be used as a platform for individual protein display. The aim of this module is to create these spores that display fusion proteins on their surface. There are several different proteins forming the spore coat layers of ''Bacillus subtilis'' spores. The outermost layer, the so called spore crust, is composed of two proteins, CotZ and CgeA ([http://www.ncbi.nlm.nih.gov/pubmed?term=imamura%20et%20al.%202011%20spore%20crust Imamura et al., 2011]). This is why we used them to create functional fusion proteins to be expressed on our '''Sporo'''beads.</p>
| + | |<p align="justify">Introduction to ''B. subtilis'' spores and their use in our project</p> |
| - | | + | |[[File:Imamura, 2011 & McKenney, 2010.png|200px|right|link=Team:LMU-Munich/Spore_Coat_Proteins/Background]] |
| - | | + | |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:Imamura, 2011 & McKenney, 2010.png|Protein distribution in spore coat of ''Bacillus subtilis''|500px|center]] | + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/Background]] |
| - | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2">Fig. 1: Composition of the ''Bacillus subtilis'' spore coat ([http://www.ncbi.nlm.nih.gov/pubmed?term=mckenney%202010%20spore%20crust McKenney ''et al''., 2010] & [http://www.ncbi.nlm.nih.gov/pubmed?term=imamura%20et%20al.%202011%20spore%20crust Imamura ''et al''., 2011])
| + | |
| - | </font>
| + | |
| - | |}
| + | |
| - | |}
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | | + | <div class="box"> |
| - | <p align="justify">The gene ''cgeA'' is located in the ''cgeABCDE'' cluster and is regulated by its own promoter P<sub>''cgeA''</sub>. The cluster ''cotVWXYZ'' contains the gene ''cotZ'' which is cotranscribed with ''cotY'' and regulated by the promoter P<sub>''cotYZ''</sub>. Another promoter of this cluster P<sub>''cotV''</sub> is responsible for the transcription of the other three genes. Those three promoters were [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters evaluated] with the ''lux'' reporter genes to get an impression of their strength and their time of activation (see for more details [http://partsregistry.org/Part:BBa_K823025 pSB<sub>''Bs''</sub>3C-''lux''ABCDE]) so they could be used for expression of spore crust fusion proteins.</p> | + | ===Cloning Strategy=== |
| - | | + | {| "width=100%" style="text-align:center;" style="align:right"| |
| - | | + | |<p align="justify">Cloning strategy to create different variants of our ''' Sporo'''beads</p> |
| - | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |[[File:Final construct.png|200px|link=Team:LMU-Munich/Spore_Coat_Proteins/cloning]] |
| - | | style="width: 100%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:Operons.png|610px|center]] | + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/cloning]] |
| - | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2">Fig. 2: Gene clusters of ''cotZ'' and ''cgeA''</font>
| + | |
| - | |}
| + | |
| - | |}
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | | + | <div class="box"> |
| - | <p align="justify">First, we used [http://partsregistry.org/Part:BBa_K823039 ''gfp''] as a proof of principle and fused it to ''cgeA'' and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 ''cotZ'']. This way we would determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.</p> | + | ===GFP as a Proof of Principle=== |
| - | | + | {| "width=100%" style="text-align:center;" style="align:right"| |
| - | | + | |<p align="justify">Main results of the various constructs that were created to find the best one!</p> |
| - | [[File:Spore crust proteins cycle.jpg|500px|center]]
| + | |[[File:LMU Firstspore.jpg|200px|right|link=Team:LMU-Munich/Spore_Coat_Proteins/result]] |
| - |
| + | |
| - | | + | |
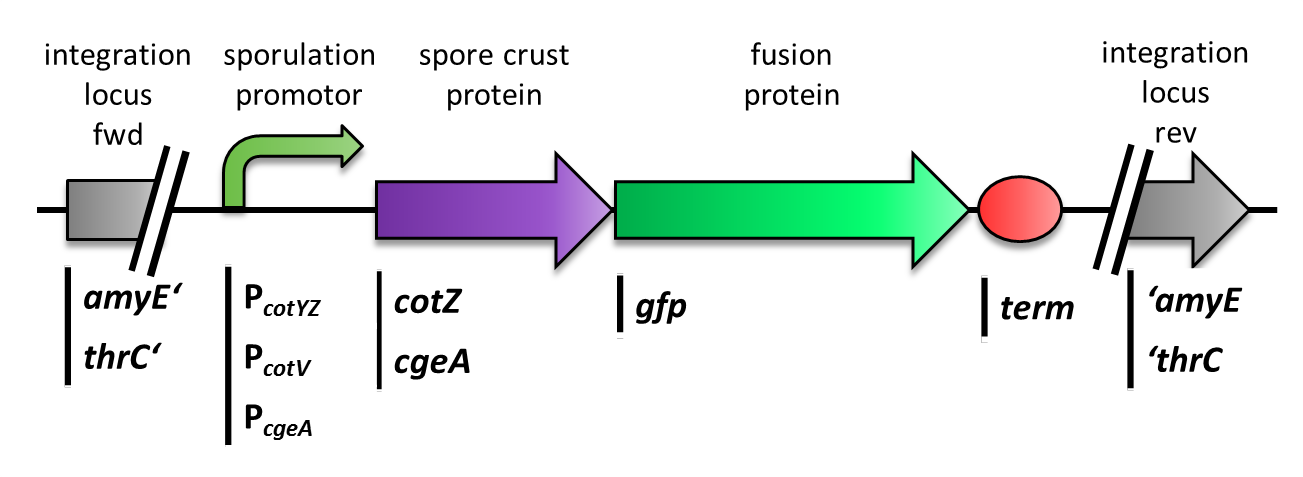
| - | <p align="justify">We constructed the BioBrick for ''cotZ'', ''cgeA'' and ''gfp'' in [http://partsregistry.org/Help:Assembly_standard_25 Freiburg Standard]. ''cotZ''was then fused to its two native promoters, P<sub>''cotV''</sub> and to P<sub>''cotYZ''</sub>, and P<sub>''cgeA''</sub>, which regulates the transcription of ''cgeA''. For ''cgeA'' we only used its native promoter P<sub>''cgeA''</sub> and the stronger one of the two promoters of the ''cotVWXYZ'' cluster, P<sub>''cotYZ''</sub> (for more details see [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters crust promotor evaluation]. While [http://partsregistry.org/Part:BBa_K823039 ''gfp''] was ligated to the terminator B0014 (see [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 Registry]). When these constructs were finished and confirmed by sequencing, we fused them together by applying the Freiburg standard to create constructs, in which gfp is fused C-terminally to ''cotZ'' or ''cgeA'' flanked by one of the three promoters and the terminator. This way we created C-terminal fusion proteins.
| + | |
| - | <br>But as we did not know if C- or N-terminal fusion would influence the fusion protein expression, our second aim was to construct N-terminal fusion proteins as well. For this purpose we wanted to fuse the genes for the crust proteins ''cotZ'' and ''cgeA'' to the terminator and ''gfp'' to the three chosen promoters. Unfortunately, there occured a mutation in the XbaI site during construction of ''gfp'' in Freiburg Standard. Therefore we were not able to finish these constructs.
| + | |
| - | </p>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:Final construct.png|610px|center]] | + | |
| - | |}
| + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/result]] |
| - | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2">Fig. 3: Section of the genome of ''B. subtilis'' with the various integrated constructs.</font>
| + | |
| - | |}
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | | + | <div class="box"> |
| - | <p align="justify">Finally we needed to clone our constructs into an empty ''Bacillus'' vector, so that they could get integrated into the genome of ''B. subtilis'' after transformation. Thus we picked the empty vector pSB<sub>BS</sub>1C from our [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors '''''Bacillus''B'''io'''B'''rick'''B'''ox], for the ''cotZ'' constructs. This vector integrates into the ''amyE'' locus, which allows to easily check the integration via starch test. In oder to also express both crust protein constructs in one strain, the ''cgeA'' fusion proteins had to be cloned into one of our other empty vectors pSB<sub>BS</sub>4S. Unfortunately for unknown reasons, the cloning of the constructs with ''cgeA'' into this vector has so far not been successful. | + | ===Laccases as functional enzymes=== |
| - | <br>
| + | {| "width=100%" style="text-align:center;" style="align:right"| |
| - | <br>We were able to finish five constructs cloned into wildtype W168 and the Δ''cotZ'' mutant:
| + | |<p align="justify">Creation of functional Laccase-''' Sporo'''beads</p> |
| - | </p>
| + | |[[File:Final construct.png|200px|link=Team:LMU-Munich/Spore_Coat_Proteins/laccases]] |
| - | | + | |
| - | | + | |
| - | {| class="colored" width="100%" align="center" style="text-align:center;" | + | |
| - | |-
| + | |
| - | |style="width:52%;"|
| + | |
| - | |style="width:20%;"| | + | |
| - | |style="width:28%;"|
| + | |
| - | |-
| + | |
| - | !
| + | |
| - | !recipient strain W168
| + | |
| - | !recipient strain W168 Δ''cotZ'' (B 49)
| + | |
| - | |-
| + | |
| - | !pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''-2aa-''gfp''-terminator
| + | |
| - | |B 53
| + | |
| - | |B 70
| + | |
| - | |-
| + | |
| - | !pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator
| + | |
| - | |B 54 | + | |
| - | |B 71 | + | |
| - | |- | + | |
| - | !pSB<sub>''Bs''</sub>1C-P<sub>''cotV''</sub>-''cotZ''-2aa-terminator
| + | |
| - | |B 55
| + | |
| - | |B 72
| + | |
| - | |-
| + | |
| - | ! pSB<sub>''Bs''</sub>1C-P<sub>''cotV''</sub>-''cotZ''-terminator
| + | |
| - | |B 56
| + | |
| - | |B 73
| + | |
| - | |-
| + | |
| - | !pSB<sub>''Bs''</sub>1C-P<sub>''cgeA''</sub>-''cotZ''-2aa-terminator
| + | |
| - | |B 52
| + | |
| - | |B 69
| + | |
| | |- | | |- |
| | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/laccases]] |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | | + | <div class="box"> |
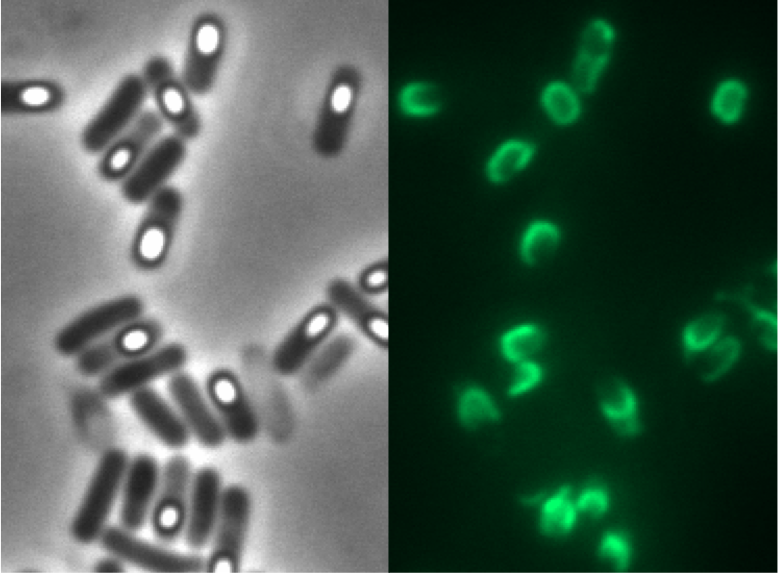
| - | <p align="justify">Finally, we started with the most important experiment for our GFP-'''Sporo'''beads, the fluorescence microscopy. Therefore we developed a sporulation protocol (for details see [https://static.igem.org/mediawiki/2012/e/e9/LMU-Munich_2012_Protocol_for_enhancement_of_mature_spore_numbers.pdf Protocol for enhancement of mature spore numbers]), that increases the rates of mature spores in our mutant samples. The cells were fixed on agarose-pads and imaged in bright field and excited in blue wavelength. All '''Sporo'''beads showed green fluorescence on their surface. But B53-'''Sporo'''bead (containing the P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator construct) illusidated the highest fluorescence intensity (see Figure 4). For further experiments, we chose this as it showed the brightest fluorescence.</p> | + | ===Purification Methods=== |
| - | | + | {| "width=100%" style="text-align:center;" style="align:right"| |
| - | | + | |<p align="justify">Description of the different purification methods of the spores</p> |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |[[File:Treatments.png|200px|right|link=Team:LMU-Munich/Spore_Coat_Proteins/purification]] |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:WT-B53 fluorescence.png|300px|center]] | + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/purification]] |
| - | {| style="color:black;" cellpadding="0" width="90%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 4: Fluorescence of wildtype spores and B53-'''Sporo'''beads (containing the P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator construct)</p></font>
| + | |
| | |} | | |} |
| - | |}
| + | </div> |
| - | |}
| + | <br> |
| - | | + | <br> |
| - | | + | <br> |
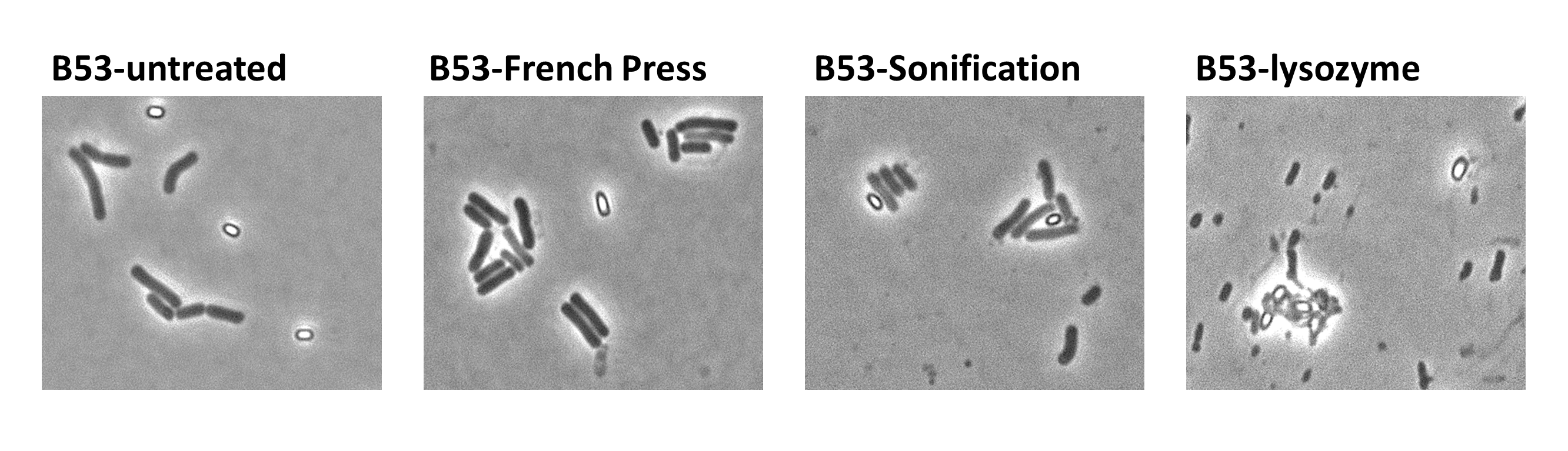
| - | <p align="justify">Since there were still some vegetative cells left after 24 hours of growth in Difco Sporulation Medium, we wanted to purify the '''Sporo'''beads. We tried three different methods for this approach: treatment with French Press, sonification and lysozyme. By means of the microscopy results we were able to conclude that lysozyme treatment was the only successful method (see [https://2012.igem.org/Team:LMU-Munich/Data/Sporepurification data]). Additionally, it did not harm the crust fusion proteins as green fluorescence was still detectable afterwards.</p>
| + | <br> |
| - | | + | <br> |
| - | <p align="justify">Eventually, clean deletions of the native genes should reveal if there is any difference in fusion protein expression in our '''Sporo'''beads. Thus we deleted the native ''cotZ'' and ''cgeA'' using the cloning method described by [http://www.ncbi.nlm.nih.gov/pubmedterm=New%20Vector%20for%20Efficient%20Allelic%20Replacement%20in%20Naturally%20Nontransformable%2C%20Low-GC-Content%2C%20Gram-Positive%20Bacteria Arnaud ''et al''., 2004].</p> | + | |
| - | | + | |
| - | | + | |
| - | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;"
| + | |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:Final construct 2.png|610px|center]]
| + | |
| - | |}
| + | |
| - | |-
| + | |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 5: Section of the genome of ''B. subtilis'' with the various integrated constructs and the deletion of ''cotZ''</p></font> | + | |
| - | |}
| + | |
| - | |}
| + | |
| - | | + | |
| - | | + | |
| - | <p align="justify">Because of the low but distinct fluorescence of wildtype spores, we measured and compared the fluorescence intensity of 100 spores per construct (see [https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore data]). The '''Sporo'''beads were investigated by fluorescence microscopy and analysed. We obtained significant differences between wildtype spores and all our '''Sporo'''beads (see [https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore Data]). The intensity bar charts should thereby show the fluorescence difference between wildtype (W168), B53- and B70-'''Sporo'''beads (B53 strain with native ''cotZ'' deletion). To demonstrate the distribution of the fusion proteins we created 3D graphs, which show the fluorescence intensity spread across the spore surface. For analysis we measured the fluorescence intensity of an area of 750px per spore by using ImageJ and evaluated it with the statistical software '''R'''. The following graph (Fig. 6) shows the results of microscopy and ImageJ analysis of the strongest construct integrated into wildtype W168 (B53) and the clean deletion mutant of ''cotZ'' (B 70).</p> | + | |
| - | | + | |
| - | | + | |
| - | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;"
| + | |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:Fluorescence of Sporobeads.png|610px|center]]
| + | |
| - | |}
| + | |
| - | |-
| + | |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | <font color="#000000"; size="2">Fig. 6: Result of fluorescence evaluation of the three strains: W168, B53 and B70.</font> | + | |
| - | |}
| + | |
| - | |}
| + | |
| - | | + | |
| - | <p align="justify">In fig. 6 it is noticable that the wildtype spore has hardly any fluorescence output whereas both spores with the integrated construct pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''-2aa-gfp-terminator give a distinct fluorescence signal. Furthermore it is shown that the strain B 70 has the highest fluorescence intesity.</p> | + | |
| - | | + | |
| - | | + | |
| | <div class="box"> | | <div class="box"> |
| | ====Project Navigation==== | | ====Project Navigation==== |
| Line 182: |
Line 81: |
| | |} | | |} |
| | </div> | | </div> |
| | + | |
| | + | |
| | | | |
| | <!-- Include the next line at the end of every page --> | | <!-- Include the next line at the end of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Footer}} | | {{:Team:LMU-Munich/Templates/Page Footer}} |







 "
"