Team:Potsdam Bioware/Biobricks/All Biobricks
From 2012.igem.org
(→Characterization:) |
(→Characterization:) |
||
| (63 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class="box_round gradient_grey"> | <div class="box_round gradient_grey"> | ||
| - | ==All | + | ==All BioBricks== |
| - | As the project is divided into three subproject, the resulting biobricks are listet & characterized in the three categories. Furthermore the Biobricks for the new RFC "Potsdam Standard" are listed.<br> | + | As the project is divided into three subproject, the resulting biobricks are listet & characterized in the three categories. Furthermore, the Biobricks for the new RFC "Potsdam Standard" are listed.<br> |
'''For detailed information on parts design, sequence, composites click the BioBrick numbers''' | '''For detailed information on parts design, sequence, composites click the BioBrick numbers''' | ||
| - | ===Antibody | + | ===Antibody BioBricks=== |
All antibody module BioBricks for the smaller construct were designed out of already existing parts and BioBricks and were expanded with further functional units by assembly PCR. The advanced antibody construct was derived by de novo synthesis based on sequence information provided by GenBank and UniProt and by one already existing Biobrick part. The gene synthesis construct was optimized for expression in CHO cell and used as template for all subsequently derived BioBricks by PCR. | All antibody module BioBricks for the smaller construct were designed out of already existing parts and BioBricks and were expanded with further functional units by assembly PCR. The advanced antibody construct was derived by de novo synthesis based on sequence information provided by GenBank and UniProt and by one already existing Biobrick part. The gene synthesis construct was optimized for expression in CHO cell and used as template for all subsequently derived BioBricks by PCR. | ||
<table border=1> | <table border=1> | ||
| Line 17: | Line 17: | ||
<tr><td>[http://partsregistry.org/Part:BBa_K929101 BBa_K929101] Anti-EGFR scFv425 with TEV site, transmembrane region an eYFP </td> | <tr><td>[http://partsregistry.org/Part:BBa_K929101 BBa_K929101] Anti-EGFR scFv425 with TEV site, transmembrane region an eYFP </td> | ||
<td> | <td> | ||
| - | The | + | The BioBrick BBa_K929101 is an extended version of the BioBrick BBa_K929100 and is composed of the already existing human EGFR (ErbB-1) signal peptide (BBa_K157001), the scFv anti-EGFR-domain-3, a TEV recognition site (according to life technologies AcTEV TM Protease manual), the transmembrane domain: transmembrane domain of B-cell receptor with glycine-serine linker (BBa_K157010) and the enhanced YFP ( BBa_E0030) reporter. The composite part was derived by PCR assembly. |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 53: | Line 53: | ||
====Characterization:==== | ====Characterization:==== | ||
=====Expression of Smaller Construct in CHO Cells===== | =====Expression of Smaller Construct in CHO Cells===== | ||
| - | |||
| + | [[Image:UP12_CHO_stabletransfected_clon.jpg|right|200px|thumb|'''Figure 1:''' eYFP fluorescence of the small construct in stably transfected CHO cells]] | ||
| + | The small antibody construct was designed with a eYFP reporter to check the a) expression level of the construct, its b) cellular localization and to enable the selection of the transfected cells via FACS. | ||
| + | The fluorescence microscopy image (Figure 1) shows the transfected CHO cells with the smaller antibody construct containing the scFV425 (BBa_K929101). It can be proven that the construct is expressed in CHO cells and also shows a high fluorescent signal. Its accumulation in the vesicular compartment of the golgi apparatus implements a membranous localization. FACS selection was successfully realized.<br><br><br><br><br><br> | ||
| + | =====Expression of Advanced Construct in CHO Cells===== | ||
| + | [[Image:UP12_CHO_stabletransfected_Big_Konstruct.jpg|right|200px|thumb|'''Figure 2:''' mCherry fluorescence of advanced antibody construct showing membrane localisation on CHO cells]] The advanced antibody construct was designed with a mCherry reporter to check the a) expression level of the construct, its b) cellular localization and to enable the selection of the transfected cells via FACS. | ||
| - | 2. | + | The fluorescence microscopy image (Figure 2) shows the transfected CHO cells with the advanced antibody construct containing the anti-GFP nanobody (PDB: 3OGO). The images indicate that the construct is expressed in CHO cells and shows a high fluorescent signal sufficient for detection. With fluorescence microscopy, confocal microscopy, immunfluorescence and FACS we were able to show the expression of the transfected parts. Under certain conditions we have seen a membrane localization of the advanced construct (Figure 2) yet we have not been able to generate cells that transport enough molecules to their surface for successful detection.<br><br><br><br><br><br><br> |
| - | ===Mutation | + | =====Functionality of Applied Cre/LoxP System===== |
| - | All the Mutation | + | [[Image:UP12_Cre_Recombinase_Blot_ohneBildunterschrift.png|right|150px|thumb|'''Figure 3:''' Western Blot of purified Nanobody-Fc from supernatant of cells co-transfected with Cre recombinase and advanced construct]] |
| + | As an additional functional unit the advanced construct contains two LoxP sites which flank the membrane anchoring region, thus allowing the switch from surface presentation to secretion on genetically level. | ||
| + | |||
| + | By co-transfecting CHO cells with our advanced antibody construct and the cre recombinase (pBS185) we were able to proof the functionality of the applied cre/LoxP system. The immune blot image (Figure 3) shows purified Nanobody-Fc from supernatant obtained from co-transfected CHO cells colored with anti-human Fc coupled to streptavidin. Purification was carried out with magnetic beads. A correct band can be seen at 41kDa.<br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | ===Mutation BioBricks=== | ||
| + | All the Mutation BioBricks are improved/modified versions of the wildtype AID that was available as a BioBrick ([http://partsregistry.org/Part:BBa_K103001 BBa_K103001]). AID is known to be responsible for somatic hypermutation and the class-switch recombination of immunoglobulin in B cells. This enzyme of 28 kDa originally occurs in B cells but does also show activity after transfection into CHO cells. AID induces the deamination of cytidine to uridine at actively transcribed single strand DNA. The replacement of cytidine by uridine leads to a mismatch during DNA replication and integrates a single base substitution predominantly in the immunoglobulin genes.<br> | ||
Therefore this enzymes was used in our project for diversification of antibodies.With PCR and cloning we developed AID`s for the differnt requirements. | Therefore this enzymes was used in our project for diversification of antibodies.With PCR and cloning we developed AID`s for the differnt requirements. | ||
| Line 95: | Line 105: | ||
====Characterization:==== | ====Characterization:==== | ||
'''Mutation rate''' in CHO-cells:<br> | '''Mutation rate''' in CHO-cells:<br> | ||
| - | [[Image:UP_12_mutation_rate.png|right|400px|thumb| | + | [[Image:UP_12_mutation_rate.png|right|400px|thumb|'''Figure 4:''' Comparison of the mutation rates between the wildtype AID, modified AID and modified AID-eGFP]] |
| - | We checked the mutation rate of wildtype AID([http://partsregistry.org/part:BBa_K929000 BBa_K929000]), modified AID([http://partsregistry.org/part:BBa_K929002 BBa_K929002]) and modified AID+eGFP([http://partsregistry.org/part:BBa_K929003 BBa_K929003]) (all expressed with CMV-promoter and hGHpolyA). Therefor we cotransfected CHO cells with a single chain construct and one of the AID versions. After certain expression time we purified the the single chain plasmids and transformed them into E.coli to enrich the mutated plasmids. After overnight culture and purification of the transformed plasmids, samples where sequenced. <br> | + | We checked the mutation rate of wildtype AID([http://partsregistry.org/part:BBa_K929000 BBa_K929000]), modified AID([http://partsregistry.org/part:BBa_K929002 BBa_K929002]) and modified AID+eGFP([http://partsregistry.org/part:BBa_K929003 BBa_K929003]) (all expressed with CMV-promoter and hGHpolyA). Therefor we cotransfected CHO cells with a single chain construct and one of the AID versions. After certain expression time we purified the the single chain plasmids and transformed them into ''E. coli'' to enrich the mutated plasmids. After overnight culture and purification of the transformed plasmids, samples where sequenced. <br> |
| - | There is a significant higher mutation rate when wildtype AID, modified AID or modified AID-eGFP is added, compared to the same experiment without AID. Surprisingly, the mutation rate of wt AID is two times higher than the mutation rate of the modified AID or modified AID-eGFP ( | + | There is a significant higher mutation rate when wildtype AID, modified AID or modified AID-eGFP is added, compared to the same experiment without AID. Surprisingly, the mutation rate of wt AID is two times higher than the mutation rate of the modified AID or modified AID-eGFP (Figure 4). This observation is contrary to our expectation. The modified AID+eGFP is located in the nucleus (Figure 5), that means our construct works and should have a higher mutation rate. One possible explanation is that the modified AID has a very high mutation rate and therefore the transfected cells die or inactivate the plasmid like it was observed for the wildtype AID (Martin and Scharff (2002)).<br><br><br> |
'''Green fluorescence reporter''':<br> | '''Green fluorescence reporter''':<br> | ||
| - | The modified AID was fused with eGFP to check 1.)the tranfection success 2.)the cellular lokalization of the improved AID and 3.) to select transfected cells via FACS. | + | The modified AID was fused with eGFP to check 1.)the tranfection success 2.)the cellular lokalization of the improved AID and 3.) to select transfected cells via FACS. <br> |
| - | + | ''' Transfection success and cellular localization: ''' | |
| - | [[Image:UP_12_green_AID_red_nanobodynew.png|right|300px|thumb| | + | [[Image:UP_12_green_AID_red_nanobodynew.png|right|300px|thumb|'''Figure 5:''' CHO cells were transfected A)with "modified AID+eGFP" (BBa_K929003)and B)with "modified AID+eGFP" (BBa_K929003) and a mCherry labeled nanobody construct (BBa_K929107)]]. |
| - | The fluorescence microscope images( | + | The fluorescence microscope images(Figure 5) show transfected cells A)with "modified AID+eGFP" (BBa_K929003)and B)with "modified AID+eGFP" (BBa_K929003) and a mCherry labeled nanobody construct (BBa_K929107)]]. "Modified AID+eGFP" (BBa_K929003)proved to be located in the nucleus. This shows that the deletion of the NES(Nuclear Export Sequence) and the addition of an NLS(Nuclear Localization Sequence) leads to an accumulation in the nucleus as it was planed. Therefore we would expect an increased mutation rate with "modified AID+eGFP" but as described above the experimental setup possible can not reflect this. It is also shown that transfected cells(green nucleus, Figure 5 A&B) can be discriminated from not transfected (grey, Figure 5). This can be used for a fast selection of transfected cells via FACS. Figure 5 B shows that "modified AID+eGFP" can be used in combination with an other fluorescence labeld construct (mCherry labeled nanobody construct ,BBa_K929107) and co transfected cells can be easily found or selected via FACS. <br><br><br><br><br><br><br><br> |
| - | === | + | ===Selection BioBricks=== |
<table border=1> | <table border=1> | ||
| - | <tr><td>[http://partsregistry.org/Part: | + | <tr><td>[http://partsregistry.org/Part:BBa_K929200 BBa_K929200] N-terminal sortase motif linked to AAV2-VP2 with Myc-tag, kozak and CMV </td> |
<td> | <td> | ||
| - | ... | + | N-terminal sortase motif is required for being ligate to several Proteins with the C-terminal sortase motif. The sortase motif was fused to the N-terminus of VP2 cap-gene, which present the fused proteins on the surface of the virus and is not essential for viral infectivity. For stronger Expression an Kozak Sequence was added. The CMV promotor is used for a high expression of the transgene. Myc-tag is used to recognize that the sortase motif is on the surface of the virus |
</td> | </td> | ||
</tr> | </tr> | ||
| - | <tr><td>[http://partsregistry.org/Part: | + | <tr><td>[http://partsregistry.org/Part:BBa_K929201 BBa_K929201] N-terminal sortase motif linked to AAV2-VP2 with kozak and CMV</td> |
<td> | <td> | ||
| - | + | The part exists of the same parts like BBa_K929200, but it lacks the myc-tag | |
</td> | </td> | ||
</tr> | </tr> | ||
| - | <tr><td>[http://partsregistry.org/Part: | + | <tr><td>[http://partsregistry.org/Part:BBa_K929202 BBa_K929202] EGF-Receptor_3rd domain with PelB leadersequence, His-tag and C-terminal Sortase motif</td> |
<td> | <td> | ||
| - | ... | + | The C-terminal sortase motif is recognized by the sortase and is cleaved between Gly and Thr. The 3rd domain of EGF-Receptor was used to target the adeno-associated virus to a high affine anti-EGFR antibody.PelB leader directs the expressed EGFR to the ''E. coli'' periplasm and the his-tag is used to purify the expressed Protein EGFR in ''E. coli''. |
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| + | ====Characterization:==== | ||
| + | [[File:UP12_mainEGFR.PNG|100px|right|thumb|'''Figure 6:''' expression of the 3rd EGFR domain in ''E. coli'' detection was performed using SDS-PAGE and Western-blot ]] | ||
| + | Using the Biobricks with the Sortase motif fused to the VP2/3 region [http://partsregistry.org/Part:BBa_K929200 BBa_K929200]; [http://partsregistry.org/Part:BBa_K929200 BBa_K929200] it is easily to ligate any proteins containing the C-terminal sortase motif with the enzyme sortase. We were interested to bring EGFR on the surface of the virus. Therefore we expressed the 3rd EGFR domain with the C-terminal sortase motif [http://partsregistry.org/Part:BBa_K929202 BBa_K929202] in ''E. coli''. We detect successful expression of EGFR on western blot (Figure 6) | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | ===Potsdam Standard BioBricks=== | ||
| + | The Potsdam Assembly Standard is a modified PLICing (Phosphorothioate-based ligase-independent gene cloning) method (Blanusa ''et al.'' (2010), Liu XP ''et al.'' (2010)). The original PLICing is based on the amplification of insert and backbone to ligate them together by knocking out thiophosphates at the 3' end. For the Potsdam Standard, we designed a new cloning vector with an RFP expression cassette as insert and two new restriction enzyme recognition sites in the suffix and prefix in pSB1C3. In the prefix we added the Apa I recognition site and in the suffix the Sph I recognition site. Both enzymes causing a 3’ overhang with 4 nucleotides. For the cloning process we cut the vector with Apa I and Sph I. In this assembly standard that’s the only step where we have to use restriction enzymes.<br> | ||
| + | The insert was amplified with primers that contain 4 phosphothioate nucleotides and the recognition sites for Apa I and Sph I at the 5’ end. After incubation in an iodine/ethanol solution the thiophosphates were cut out resulting in a 3’ overhang which is suitable to the overhangs that was created by cutting the new assembly vector. | ||
| + | After that the digested vector and the PLICed insert were mixed and transformed into <i>E. coli</i>. By using the RFP expression cassette we created a ligation control system. Due to the fact that red fluorescent colonies have a failed vector ligation we can tell which colonies are correctly ligated. The colonies that do not show red fluorescence are the positive clones. | ||
| + | |||
| + | <table border=1> | ||
| + | <tr><td>[http://partsregistry.org/Part:BBa_K929301 BBa_K929301] Backbone of the Potsdam Standard </td> | ||
| + | <td> | ||
| + | backbone contains two new restriction sites: Apa I and Sph I and a RFP expression cassette as ligation control | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr><td>[http://partsregistry.org/Part:BBa_K929302 BBa_K929302] AID in Potsdam Standard</td> | ||
| + | <td> | ||
| + | AID in new Potsdam Standard vector with inactivated Apa I and Sph I, but active RFC 10 restriction sites | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | ====Characterization:==== | ||
| + | <br> | ||
| + | We tested different combination to prove the most efficient experimental design. Therefore, we tried four different conditions: the ligation of the pliced GOI with the unpurified digested backbone with and without ligase and the ligation of the pliced GOI with the purified digested backbone with and without ligase. After transformation, we picked only clones which have lost their red fluorescence indicating that the ligation was successful (Figure 9).<br> | ||
| + | To quantify the results, we counted the red colonies and the colonies without red fluorescence (Figure 10). It is obvious, that the ligation with the electrophoretic purified digested backbone is more efficient than without any purification. Nevertheless, the use of ligase is not necessary for the ligation process under these conditions.<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <table class="table center" border=0><tr><td>[[file:RFP_culture.jpg|400px|center|thumb|'''Figure 7:''' Overnight culture of transformed ''E. coli'']]</td><td>[[File:UP12_RFP_pellets.jpg|400px|center|thumb|'''Figure 8:''' After centrifugation, the cell pellets are coloured like cherries]]</td><tr><td>[[file:UP12_Ligation.png|400px|center|thumb|'''Figure 9:''' Results of different ligation conditions]]</td><td>[[file:UP_12_Potstdam_Standard.png|400px|center|thumb|'''Figure 10:''' Results of different ligation conditions]]</td></tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | </div> | ||
{{:Team:Potsdam_Bioware/css}} | {{:Team:Potsdam_Bioware/css}} | ||
Latest revision as of 17:11, 26 October 2012
Contents |
All BioBricks
As the project is divided into three subproject, the resulting biobricks are listet & characterized in the three categories. Furthermore, the Biobricks for the new RFC "Potsdam Standard" are listed.
For detailed information on parts design, sequence, composites click the BioBrick numbers
Antibody BioBricks
All antibody module BioBricks for the smaller construct were designed out of already existing parts and BioBricks and were expanded with further functional units by assembly PCR. The advanced antibody construct was derived by de novo synthesis based on sequence information provided by GenBank and UniProt and by one already existing Biobrick part. The gene synthesis construct was optimized for expression in CHO cell and used as template for all subsequently derived BioBricks by PCR.
| [http://partsregistry.org/Part:BBa_K929100 BBa_K929100] Anti-EGFR scFv425 |
The single chain fragment (scFV425) specifically binds to the epidermal growth factor receptor (EGFR) domain 3. The part has a shortened N-terminal FLAG-tag sequence of five amino acids (DYKDE) for purification and detection. |
| [http://partsregistry.org/Part:BBa_K929101 BBa_K929101] Anti-EGFR scFv425 with TEV site, transmembrane region an eYFP |
The BioBrick BBa_K929101 is an extended version of the BioBrick BBa_K929100 and is composed of the already existing human EGFR (ErbB-1) signal peptide (BBa_K157001), the scFv anti-EGFR-domain-3, a TEV recognition site (according to life technologies AcTEV TM Protease manual), the transmembrane domain: transmembrane domain of B-cell receptor with glycine-serine linker (BBa_K157010) and the enhanced YFP ( BBa_E0030) reporter. The composite part was derived by PCR assembly. |
| [http://partsregistry.org/Part:BBa_K929102 BBa_K929102] Anti-GFP Nanobody with IgG kappa signal peptide and IgG1 Fc domain |
This part represents an antibody unit composed of a IgG kappa signal peptide (UniProt: P01601), an anti-GFP nanobody (PDB: 3OGO) and an IgG1 Fc domain (UniProt: P01857). |
| [http://partsregistry.org/Part:BBa_K929103 BBa_K929103] B-cell transmembrane region with mCherry and TEV site, framed by LoxP |
This part represents the switchable membrane anchoring region of or antibody construct (BBa_K929107) and consists of the TEV recognition site, the modified transmembrane domain of B-cell receptor with a shortened linker (modified BBa_K157010), the mCherry reporter and two framing LoxP sites. |
| [http://partsregistry.org/Part:BBa_K929104 BBa_K929104] Anti-GFP Nanobody |
The green fluorescent protein (GFP)-nanobody is a single-chain VHH antibody domain which shows specific binding activity against GFP (PDB: 3OGO). |
| [http://partsregistry.org/Part:BBa_K929105 BBa_K929105] IgG1 Fc domain |
A human immunoglobulin gamma-1 heavy chain constant region (IGHG1) was added to the construct and enables the direct interaction of the antibody unit with the Fc receptor and complement proteins (UniProt: P01857). |
| [http://partsregistry.org/Part:BBa_K929106 BBa_K929106] B-cell transmembrane region with mCherry |
This part is a composed part of the transmembrane domain of a B-cell receptor (modified BBa_K157010)fused to the mCherry reporter for detection of cellular localization. |
| [http://partsregistry.org/Part:BBa_K929107 BBa_K929107] Anti-GFP, IgG1 Fc with TEV site, TMR, mCherry, framed by LoxP |
The antibody construct consists of two major building blocks represented by the actual antibody unit ( BBa_K929102) and the switchable membrane anchoring region (BBa_K929103). The antibody unit is represented by the human Ig kappa chain V-I region signal peptide (UniProt: P01601), the anti-GFP Nanobody (PDB: 3OGO) and the Fc region (UniProt: P01857). TEV protease recognition site, 2 LoxP sites, the B-cell receptor transmembrane domain (modified BBa_K157010) and the mCherry reporter display the switchable membrane anchoring region. |
Characterization:
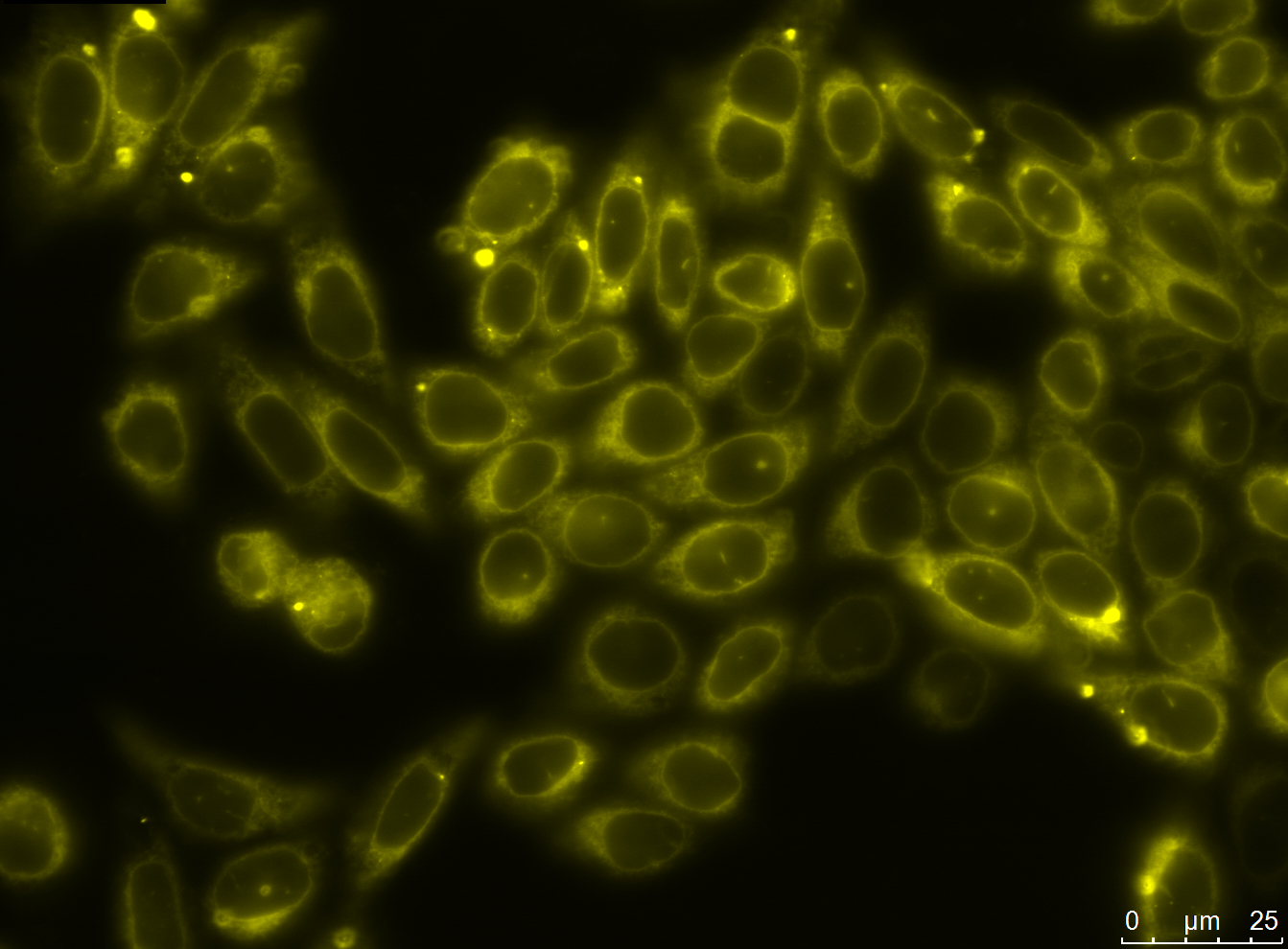
Expression of Smaller Construct in CHO Cells
The small antibody construct was designed with a eYFP reporter to check the a) expression level of the construct, its b) cellular localization and to enable the selection of the transfected cells via FACS.
The fluorescence microscopy image (Figure 1) shows the transfected CHO cells with the smaller antibody construct containing the scFV425 (BBa_K929101). It can be proven that the construct is expressed in CHO cells and also shows a high fluorescent signal. Its accumulation in the vesicular compartment of the golgi apparatus implements a membranous localization. FACS selection was successfully realized.
Expression of Advanced Construct in CHO Cells
The fluorescence microscopy image (Figure 2) shows the transfected CHO cells with the advanced antibody construct containing the anti-GFP nanobody (PDB: 3OGO). The images indicate that the construct is expressed in CHO cells and shows a high fluorescent signal sufficient for detection. With fluorescence microscopy, confocal microscopy, immunfluorescence and FACS we were able to show the expression of the transfected parts. Under certain conditions we have seen a membrane localization of the advanced construct (Figure 2) yet we have not been able to generate cells that transport enough molecules to their surface for successful detection.
Functionality of Applied Cre/LoxP System
As an additional functional unit the advanced construct contains two LoxP sites which flank the membrane anchoring region, thus allowing the switch from surface presentation to secretion on genetically level.
By co-transfecting CHO cells with our advanced antibody construct and the cre recombinase (pBS185) we were able to proof the functionality of the applied cre/LoxP system. The immune blot image (Figure 3) shows purified Nanobody-Fc from supernatant obtained from co-transfected CHO cells colored with anti-human Fc coupled to streptavidin. Purification was carried out with magnetic beads. A correct band can be seen at 41kDa.
Mutation BioBricks
All the Mutation BioBricks are improved/modified versions of the wildtype AID that was available as a BioBrick ([http://partsregistry.org/Part:BBa_K103001 BBa_K103001]). AID is known to be responsible for somatic hypermutation and the class-switch recombination of immunoglobulin in B cells. This enzyme of 28 kDa originally occurs in B cells but does also show activity after transfection into CHO cells. AID induces the deamination of cytidine to uridine at actively transcribed single strand DNA. The replacement of cytidine by uridine leads to a mismatch during DNA replication and integrates a single base substitution predominantly in the immunoglobulin genes.
Therefore this enzymes was used in our project for diversification of antibodies.With PCR and cloning we developed AID`s for the differnt requirements.
| [http://partsregistry.org/Part:BBa_K929000 BBa_K929000] AID with CMV promoter and hGH-polyadenylation signal sequence |
CMV-promoter & hGH polyadenylationsequence were added to the wildtype AID for strong expression. |
| [http://partsregistry.org/Part:BBa_K929001 BBa_K929001] modified_AID without NES, with NLS and Kozak sequence |
To improve the mutation rate the AID was located into the nucleus, because there it mutates transcribed DNA. Therefor an aditional NLS(Nuclear Localization Sequence)was added and the naturally occurring NES(Nuclear Exprot Sequence) was removed.For stronger Expression an Kozak Sequence was added. To fuse modified AID((C-terminal of mod. AID) with RFC 25 parts, we added an AgeI restriction site. |
| [http://partsregistry.org/Part:BBa_K929002 BBa_K929002] modified_AID with CMV and hGH-polyA |
For strong expression of the previously described part an CMV-promoter and hGH polyadenylation sequence were added. |
| [http://partsregistry.org/Part:BBa_K929004 BBa_K929004] modified_AID+eGFP |
The modified AID was fused with eGFP to check 1.)the tranfection success 2.)the cellular lokalization of the improved AID and 3.) to select transfected cells via FACS. |
| [http://partsregistry.org/Part:BBa_K929003 BBa_K929003]modified AID with CMV, hGH-polyA and eGFP |
For strong expression of the previously described part an CMV-promoter and hGH polyadenylation sequence were added. |
Characterization:
Mutation rate in CHO-cells:
We checked the mutation rate of wildtype AID([http://partsregistry.org/part:BBa_K929000 BBa_K929000]), modified AID([http://partsregistry.org/part:BBa_K929002 BBa_K929002]) and modified AID+eGFP([http://partsregistry.org/part:BBa_K929003 BBa_K929003]) (all expressed with CMV-promoter and hGHpolyA). Therefor we cotransfected CHO cells with a single chain construct and one of the AID versions. After certain expression time we purified the the single chain plasmids and transformed them into E. coli to enrich the mutated plasmids. After overnight culture and purification of the transformed plasmids, samples where sequenced.
There is a significant higher mutation rate when wildtype AID, modified AID or modified AID-eGFP is added, compared to the same experiment without AID. Surprisingly, the mutation rate of wt AID is two times higher than the mutation rate of the modified AID or modified AID-eGFP (Figure 4). This observation is contrary to our expectation. The modified AID+eGFP is located in the nucleus (Figure 5), that means our construct works and should have a higher mutation rate. One possible explanation is that the modified AID has a very high mutation rate and therefore the transfected cells die or inactivate the plasmid like it was observed for the wildtype AID (Martin and Scharff (2002)).
Green fluorescence reporter:
The modified AID was fused with eGFP to check 1.)the tranfection success 2.)the cellular lokalization of the improved AID and 3.) to select transfected cells via FACS.
Transfection success and cellular localization:
The fluorescence microscope images(Figure 5) show transfected cells A)with "modified AID+eGFP" (BBa_K929003)and B)with "modified AID+eGFP" (BBa_K929003) and a mCherry labeled nanobody construct (BBa_K929107)]]. "Modified AID+eGFP" (BBa_K929003)proved to be located in the nucleus. This shows that the deletion of the NES(Nuclear Export Sequence) and the addition of an NLS(Nuclear Localization Sequence) leads to an accumulation in the nucleus as it was planed. Therefore we would expect an increased mutation rate with "modified AID+eGFP" but as described above the experimental setup possible can not reflect this. It is also shown that transfected cells(green nucleus, Figure 5 A&B) can be discriminated from not transfected (grey, Figure 5). This can be used for a fast selection of transfected cells via FACS. Figure 5 B shows that "modified AID+eGFP" can be used in combination with an other fluorescence labeld construct (mCherry labeled nanobody construct ,BBa_K929107) and co transfected cells can be easily found or selected via FACS.
Selection BioBricks
| [http://partsregistry.org/Part:BBa_K929200 BBa_K929200] N-terminal sortase motif linked to AAV2-VP2 with Myc-tag, kozak and CMV |
N-terminal sortase motif is required for being ligate to several Proteins with the C-terminal sortase motif. The sortase motif was fused to the N-terminus of VP2 cap-gene, which present the fused proteins on the surface of the virus and is not essential for viral infectivity. For stronger Expression an Kozak Sequence was added. The CMV promotor is used for a high expression of the transgene. Myc-tag is used to recognize that the sortase motif is on the surface of the virus |
| [http://partsregistry.org/Part:BBa_K929201 BBa_K929201] N-terminal sortase motif linked to AAV2-VP2 with kozak and CMV |
The part exists of the same parts like BBa_K929200, but it lacks the myc-tag |
| [http://partsregistry.org/Part:BBa_K929202 BBa_K929202] EGF-Receptor_3rd domain with PelB leadersequence, His-tag and C-terminal Sortase motif |
The C-terminal sortase motif is recognized by the sortase and is cleaved between Gly and Thr. The 3rd domain of EGF-Receptor was used to target the adeno-associated virus to a high affine anti-EGFR antibody.PelB leader directs the expressed EGFR to the E. coli periplasm and the his-tag is used to purify the expressed Protein EGFR in E. coli. |
Characterization:
Using the Biobricks with the Sortase motif fused to the VP2/3 region [http://partsregistry.org/Part:BBa_K929200 BBa_K929200]; [http://partsregistry.org/Part:BBa_K929200 BBa_K929200] it is easily to ligate any proteins containing the C-terminal sortase motif with the enzyme sortase. We were interested to bring EGFR on the surface of the virus. Therefore we expressed the 3rd EGFR domain with the C-terminal sortase motif [http://partsregistry.org/Part:BBa_K929202 BBa_K929202] in E. coli. We detect successful expression of EGFR on western blot (Figure 6)
Potsdam Standard BioBricks
The Potsdam Assembly Standard is a modified PLICing (Phosphorothioate-based ligase-independent gene cloning) method (Blanusa et al. (2010), Liu XP et al. (2010)). The original PLICing is based on the amplification of insert and backbone to ligate them together by knocking out thiophosphates at the 3' end. For the Potsdam Standard, we designed a new cloning vector with an RFP expression cassette as insert and two new restriction enzyme recognition sites in the suffix and prefix in pSB1C3. In the prefix we added the Apa I recognition site and in the suffix the Sph I recognition site. Both enzymes causing a 3’ overhang with 4 nucleotides. For the cloning process we cut the vector with Apa I and Sph I. In this assembly standard that’s the only step where we have to use restriction enzymes.
The insert was amplified with primers that contain 4 phosphothioate nucleotides and the recognition sites for Apa I and Sph I at the 5’ end. After incubation in an iodine/ethanol solution the thiophosphates were cut out resulting in a 3’ overhang which is suitable to the overhangs that was created by cutting the new assembly vector.
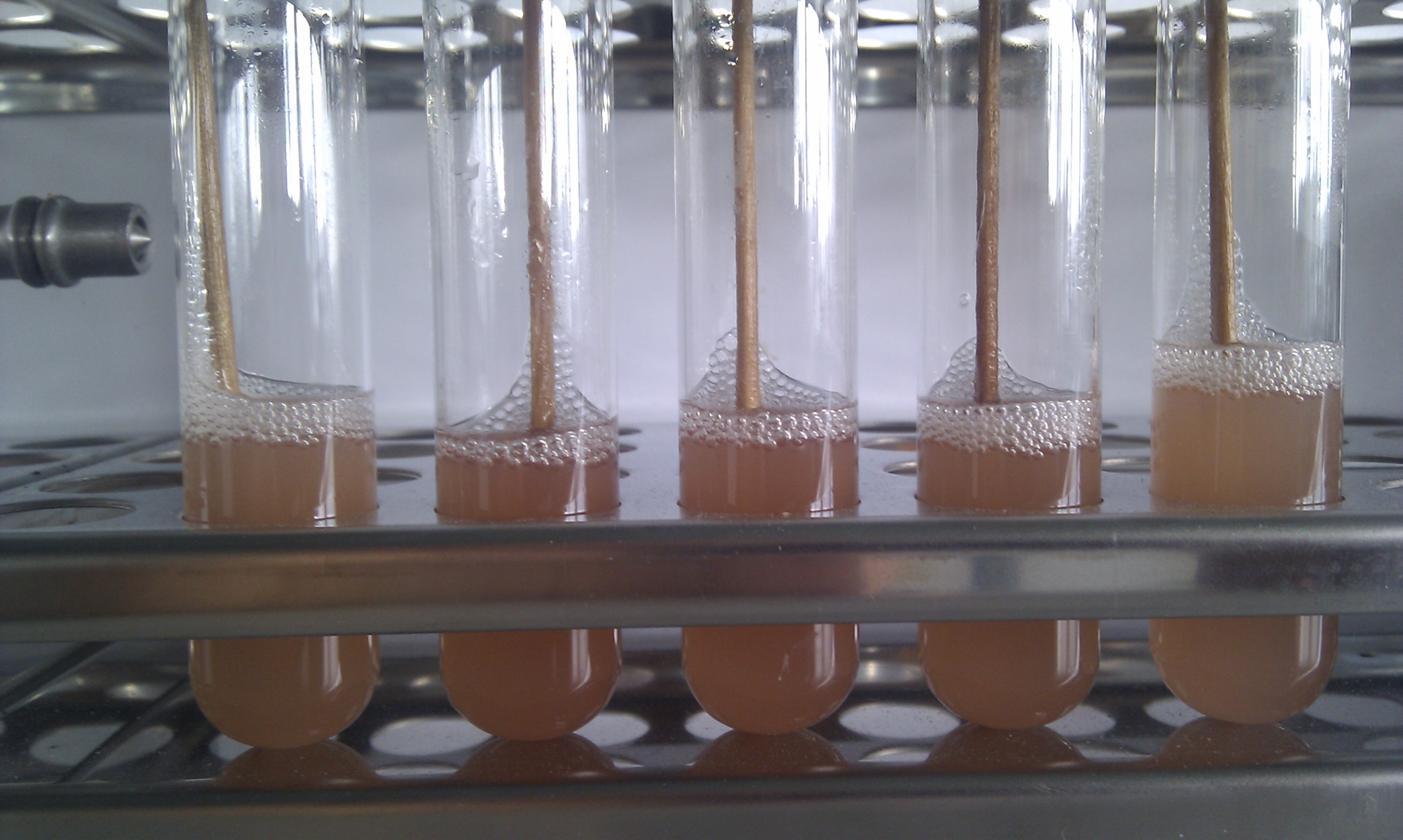
After that the digested vector and the PLICed insert were mixed and transformed into E. coli. By using the RFP expression cassette we created a ligation control system. Due to the fact that red fluorescent colonies have a failed vector ligation we can tell which colonies are correctly ligated. The colonies that do not show red fluorescence are the positive clones.
| [http://partsregistry.org/Part:BBa_K929301 BBa_K929301] Backbone of the Potsdam Standard |
backbone contains two new restriction sites: Apa I and Sph I and a RFP expression cassette as ligation control |
| [http://partsregistry.org/Part:BBa_K929302 BBa_K929302] AID in Potsdam Standard |
AID in new Potsdam Standard vector with inactivated Apa I and Sph I, but active RFC 10 restriction sites |
Characterization:
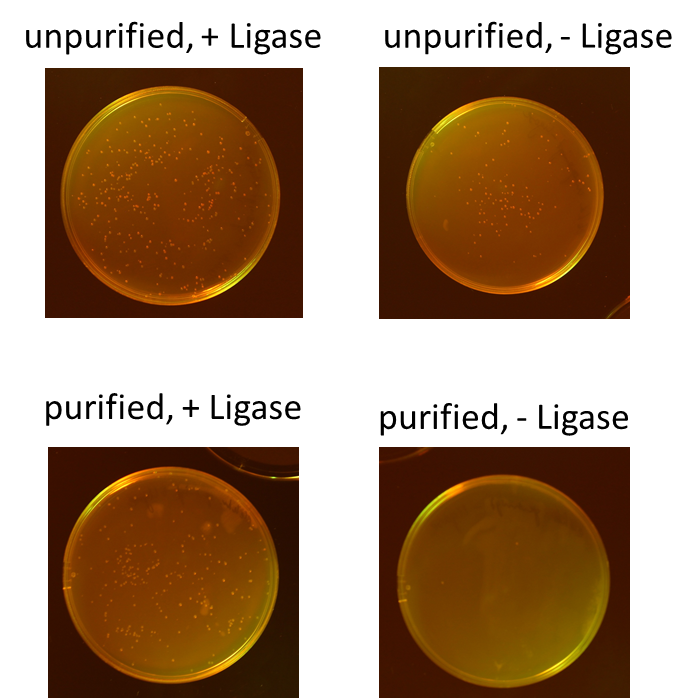
We tested different combination to prove the most efficient experimental design. Therefore, we tried four different conditions: the ligation of the pliced GOI with the unpurified digested backbone with and without ligase and the ligation of the pliced GOI with the purified digested backbone with and without ligase. After transformation, we picked only clones which have lost their red fluorescence indicating that the ligation was successful (Figure 9).
To quantify the results, we counted the red colonies and the colonies without red fluorescence (Figure 10). It is obvious, that the ligation with the electrophoretic purified digested backbone is more efficient than without any purification. Nevertheless, the use of ligase is not necessary for the ligation process under these conditions.
 "
"