Team:Wageningen UR/Journal/week17
From 2012.igem.org
(→Lab work) |
(→Lab work) |
||
| (One intermediate revision not shown) | |||
| Line 19: | Line 19: | ||
Did a colony PCR on one transformant colony (Could only pick one, because of overgrowth). | Did a colony PCR on one transformant colony (Could only pick one, because of overgrowth). | ||
<br/> | <br/> | ||
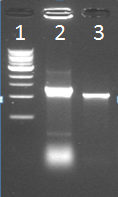
| - | [[File:21_08_colony_pcr.PNG|250px|left|thumb|Figure 1: Colony PCR of one IPTG_CCMV_NEG colony]]<br/><br/><br/><br/> | + | [[File:21_08_colony_pcr.PNG|250px|left|thumb|''Figure 1: Colony PCR of one IPTG_CCMV_NEG colony'']]<br/><br/><br/><br/> |
Lane 1: Marker<br/> | Lane 1: Marker<br/> | ||
Lane 2: Kees’s sample<br/> | Lane 2: Kees’s sample<br/> | ||
| - | Lane 3: | + | Lane 3: IPTG_CCMV_NEG in IPTG backbone (BBa_J04500) in JM 109 cells |
<br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/> | ||
| Line 49: | Line 49: | ||
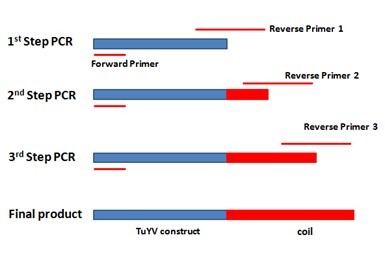
For adding the E.coil to the C terminal of monomer, three step extension PCR was designed in order to fuse the coil gene to the 3’ end of our construct gene. Specific reverse primer was designed for each step, totally three. The primer’s 5’ end contains new sequence and 3’ end will bind to the template. After each step PCR reaction, the new sequence will be added to the 3’ end of the template and the PCR products from this step will be utilized as template for the next step. Finally, the 63bp E.coil will be added to the 3’ end of our construct gene. The overall scheme is showed in the figure below. | For adding the E.coil to the C terminal of monomer, three step extension PCR was designed in order to fuse the coil gene to the 3’ end of our construct gene. Specific reverse primer was designed for each step, totally three. The primer’s 5’ end contains new sequence and 3’ end will bind to the template. After each step PCR reaction, the new sequence will be added to the 3’ end of the template and the PCR products from this step will be utilized as template for the next step. Finally, the 63bp E.coil will be added to the 3’ end of our construct gene. The overall scheme is showed in the figure below. | ||
</p> | </p> | ||
| - | [[File:the overall scheme for adding E.coil to the 3’ end of TuYV construct..jpg|500px|center|thumb|Figure | + | [[File:the overall scheme for adding E.coil to the 3’ end of TuYV construct..jpg|500px|center|thumb|''Figure 2: ...'']] |
<p align="justify"> | <p align="justify"> | ||
| Line 102: | Line 102: | ||
* repeat 3rd step of the PCR reaction with the sample of 15. Aug | * repeat 3rd step of the PCR reaction with the sample of 15. Aug | ||
| - | [[File:PCR 2nd,3rd step hepinsidecoil.png|250px|center|thumb|Figure | + | [[File:PCR 2nd,3rd step hepinsidecoil.png|250px|center|thumb|''Figure 3: 2nd and 3rd step of the HepB inside modification PCR reaction'']] |
-> 3rd step of the PCR reaction seems to work (right size + size difference compared to the 2nd step, but also a smear is visible | -> 3rd step of the PCR reaction seems to work (right size + size difference compared to the 2nd step, but also a smear is visible | ||
Latest revision as of 03:35, 27 September 2012
week 17: 20 august - 26 august
Office work
Lab work
CCMV
20th August (Hugo)
Ligated IPTG_CCMV_NEG into the IPTG backbone (BBa_J04500) and transformed into JM 109 competent cells using electrophoresis.
21st August (Hugo)
Did a colony PCR on one transformant colony (Could only pick one, because of overgrowth).
Lane 1: Marker
Lane 2: Kees’s sample
Lane 3: IPTG_CCMV_NEG in IPTG backbone (BBa_J04500) in JM 109 cells
22nd August (Hugo)
Inoculated 10 mL of LB with IPTG_CCMV_NEG JM 109 cells.
23rd August (Hugo)
did a colony PCR on iron inducible biobricks and IPTG_CCMV_NEG.
Inoculated 100 mL LB medium with 2 mL of IPTG_CCMV_NEG cell culture. Induced before OD600 was 0.6 and grew for another 4 hrs. Then pelleted the culture, followed by washing steps. Stored the pellet in the freezer overnight.
24th August (Hugo)
Disrupted the pelleted cells using a french press and prepared the solution for dialysis. Ran dialysis over the weekend.
TuYV
On the basis of successfully getting TuYV VLPs, modifications with two protein coil-structures will be explored on the outside of TUYV VLPs. The two coils, E/K.coil, have electrostatic interaction due to different charge and can serve as an anchor for other proteins. K.coil, which is positive charged, will be attached to the C terminus of the VLPs monomer and E.coil, which is negative charged, will be attached with other proteins, such as using GFP as reporter ligand.
For adding the E.coil to the C terminal of monomer, three step extension PCR was designed in order to fuse the coil gene to the 3’ end of our construct gene. Specific reverse primer was designed for each step, totally three. The primer’s 5’ end contains new sequence and 3’ end will bind to the template. After each step PCR reaction, the new sequence will be added to the 3’ end of the template and the PCR products from this step will be utilized as template for the next step. Finally, the 63bp E.coil will be added to the 3’ end of our construct gene. The overall scheme is showed in the figure below.
This week we started adding the coil to the C terminal of TuYV constructs. With designed primers, the first step PCR showed bands on the right sizes. The second step PCR was tried three times this week, but we always did not get any band on the gel check for second step PCR. This was because after the first step, we did gel extraction for our first step PCR products, but the DNA concentration was very low after gel extraction and the template was destroyed during the extraction process.
TuYV constructs in Ampicillin resistant backbone and BBa_J04500 were digested and checked on the gel, but we did not see any band on the gel check for all 4 sample. Later, we loaded the miniprep products on the gel, we got smear, it turned out to be our miniprep products were degraded.
TuYV constructs (PCR procucts: 1, 1H, 4, 4H) were digested, ligated with plasmid containing IPTG induced promoter and transformed into JM-109 again, we got very promising plate, there were lot of colonies on the sample plates.
Hepatitis B
20 August
- Colony PCR
- of HepB (without promoter) in pSB1C3 - of HepB with a his tag + IPTG promoter in Bba_J04500 -> the samples show no band on the gel - so the transformation did not succeed
- Ligation of the HepB with the IPTG promoter into pSB1C3
- Transformation of the ligation with both JM109 and DH5α; using the BBa_J04450 brick as positive control
22 August
- Colony PCR
- Of the transformations HepB + IPTGpromoter in pSB1C3 backbone in JM109 and DH5α -> the samples show no band on the gel - so the transformation did not succeed
23 August
- 2nd try - Colony PCR
Of the transformations GFP-coil in pSB1C3 backbone in JM109 and DH5α -> again no band on the gel
Hepatitis B inside modification
23 August
- Step 3 and step 4 of the PCR reaction
-> no bands visible for the 3rd and 4th step of HepBinside-coil
- control of a miniprep of HepB+IPTG in BL21 from 16.Aug (sample from 6.Aug)
-> the miniprep shows a band of the expected size - so the transformation from 6.Aug was successful
24 August
- repeat 3rd step of the PCR reaction with the sample of 15. Aug
-> 3rd step of the PCR reaction seems to work (right size + size difference compared to the 2nd step, but also a smear is visible
GFP modification
20 August
- Ligation of the digested GFP-coil PCR product from 15.Aug into BBa_J04450
- Transformation with both JM109 and DH5α; using the BBa_J04450 brick as positive control
22 August
- Colony PCR of the transformations GFP-coil in pSB1C3 backbone in JM109 and DH5α
23 August
- 2nd try - Colony PCR of the transformations GFP-coil in pSB1C3 backbone in JM109 DH5α
24 August
- check and amplify GFP-coil PCR product from 14.Aug
-> no conclusion can be made because of very weak bands
CCMV Stability experiments
20th – 21st August (Mark)
- Temperature and time stability experiment with DLS
- Temperature range 5-40 degree Celcius
- Time measuring points 3, 6 and 24 hours after start experiment
- DLS measurment: 20 runs of 35 seconds
PLRV
Placing the PLRV CP’s downstream a IPTG inducible promoter. For expression of the PLRV CP’s we inserted the CP genes downstream an IPTG inducible promoter and a ribosomal binding site (RBS). The products were used to transform Escherichia coli (E. coli) DH5α cells. After growth, induction an cell lysis (see French press protocol) we ran a SDS-PAGE gel. Unfortunately no difference in protein expression was seen.
 "
"