Team:HokkaidoU Japan/Notebook/aggregation Week 11
From 2012.igem.org
| (One intermediate revision not shown) | |||
| Line 4: | Line 4: | ||
<!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 10th== | + | ===September 10th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2== | + | ====Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2==== |
| - | + | ||
To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 by 3piece ligation, we digested pBAD-RBS-eCFP-RBS-pSB1A2 by EcoRI and SpeI, Ag43-dT-pSB1AK3 (previously digested by HindIII) by XbaI and NotI, and pSTV28 by EcoRI and NotI. Then we digested pT7-RBS-pSB1C3 by XbaI and SpeI as a control for confirmation of the ability of restriction enzyme. | To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 by 3piece ligation, we digested pBAD-RBS-eCFP-RBS-pSB1A2 by EcoRI and SpeI, Ag43-dT-pSB1AK3 (previously digested by HindIII) by XbaI and NotI, and pSTV28 by EcoRI and NotI. Then we digested pT7-RBS-pSB1C3 by XbaI and SpeI as a control for confirmation of the ability of restriction enzyme. | ||
<br /> | <br /> | ||
| Line 31: | Line 30: | ||
|30 ul | |30 ul | ||
|} | |} | ||
| - | |||
| Line 58: | Line 56: | ||
|30 ul | |30 ul | ||
|} | |} | ||
| - | |||
| - | |||
| Line 109: | Line 105: | ||
|20 ul | |20 ul | ||
|} | |} | ||
| - | |||
| Line 130: | Line 125: | ||
|HOLD | |HOLD | ||
|} | |} | ||
| - | |||
[[image:HokkaidoU2012 120910 digestion Insert1-pBAD-RBS-eCFP-RBS-pSB1A2 Insert2-Ag43-dT-pSB1AK3 Vector-ptet-RBS-eYFP-dT-pSTV28 Control-pT7-RBS-pSB1A2.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120910 digestion Insert1-pBAD-RBS-eCFP-RBS-pSB1A2 Insert2-Ag43-dT-pSB1AK3 Vector-ptet-RBS-eYFP-dT-pSTV28 Control-pT7-RBS-pSB1A2.jpg|thumb|digestion result]] | ||
| - | |||
| - | ==Ethanol precipitation of ptet-RBS-eYFP-dT-pSB1A2 and pSTV28== | + | ====Ethanol precipitation of ptet-RBS-eYFP-dT-pSB1A2 and pSTV28==== |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
#Centrifuged at 14000 rpm, 30 min at 4C. | #Centrifuged at 14000 rpm, 30 min at 4C. | ||
| Line 142: | Line 134: | ||
#Centrifuged at 15000 rpm, 15 min at 4C. | #Centrifuged at 15000 rpm, 15 min at 4C. | ||
#Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| - | |||
[[image:HokkaidoU2012 120910 EthaPre Insert1-Insert2-Vector.jpg|thumb|ethanol precipitation result]] | [[image:HokkaidoU2012 120910 EthaPre Insert1-Insert2-Vector.jpg|thumb|ethanol precipitation result]] | ||
We confirmed that the concentration of Insert1 DNA solution is 50 ng/ul, Insert2 DNA solution is 10~15 ng/ul and Vector DNA solution is 10 ng/ul. | We confirmed that the concentration of Insert1 DNA solution is 50 ng/ul, Insert2 DNA solution is 10~15 ng/ul and Vector DNA solution is 10 ng/ul. | ||
| - | |||
| - | ==Ligation of pBAD-RBS-eCFP-RBS, Ag43-dT and pSTV28== | + | ====Ligation of pBAD-RBS-eCFP-RBS, Ag43-dT and pSTV28==== |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 186: | Line 176: | ||
|} | |} | ||
| - | |||
| - | ==Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28== | + | ====Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28==== |
| - | + | ||
Transformation ligation product into DH5α. | Transformation ligation product into DH5α. | ||
#Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 ligation product to 50 ul of thawed competent cells on ice. | ||
| Line 199: | Line 187: | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
#Incubated the plates at 37C for 14 hrs. | #Incubated the plates at 37C for 14 hrs. | ||
| - | |||
| - | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | |||
| + | </div></div> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 11th== | + | ===September 11th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Colony PCR of pBAD-RBS-eCFP-RBS-pSB1A2== | + | ====Colony PCR of pBAD-RBS-eCFP-RBS-pSB1A2==== |
| - | + | ||
| Line 227: | Line 215: | ||
|10 ul | |10 ul | ||
|} | |} | ||
| - | |||
{|class="hokkaidou-table-pcr-time" | {|class="hokkaidou-table-pcr-time" | ||
| Line 267: | Line 254: | ||
[[image:HokkaidoU2012 120911 colop pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28.jpg|thumb|Colony PCR result]] | [[image:HokkaidoU2012 120911 colop pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28.jpg|thumb|Colony PCR result]] | ||
| - | |||
Because of mis-setting the PCR program, we failed to amplify desired DNA fragment so we retried the colony PCR in same reagent, correct PCR machine program. | Because of mis-setting the PCR program, we failed to amplify desired DNA fragment so we retried the colony PCR in same reagent, correct PCR machine program. | ||
| Line 273: | Line 259: | ||
[[image:HokkaidoU2012 120912 colop2 pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28.jpg|thumb|Colony PCR result]] | [[image:HokkaidoU2012 120912 colop2 pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28.jpg|thumb|Colony PCR result]] | ||
| + | We noticed that the ligated DNA contains at least Ag43-dT-BioBrick_Suffix complex. We selected No. 2,4 for incubation for plasmid extraction and No. 5,6 for Aggregation check. | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| - | |||
| - | |||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 12th== | + | ===September 12th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Analysis nucleotide sequence== | + | ====Analysis nucleotide sequence==== |
| - | + | ||
We analyzed the nucleotide sequence of pBAD-RBS-Ag43-dT on pSB1AK3 and pSB1C3. | We analyzed the nucleotide sequence of pBAD-RBS-Ag43-dT on pSB1AK3 and pSB1C3. | ||
We used these 6 kinds of primers. | We used these 6 kinds of primers. | ||
100b up EX-F primer, pBAD-f1 primer, pBAD-f2 primer, Ag43-f1 primer, Ag43-f2 primer, Ag43-f3 primer, Ag43-f4 primer, 200b down PS-R primer | 100b up EX-F primer, pBAD-f1 primer, pBAD-f2 primer, Ag43-f1 primer, Ag43-f2 primer, Ag43-f3 primer, Ag43-f4 primer, 200b down PS-R primer | ||
| - | + | ====Aggregation check of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28==== | |
| - | + | ||
| - | ==Aggregation check of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28== | + | |
| - | + | ||
Aggregation check for No. 5,6 colonies selected by colony PCR at August 11th. | Aggregation check for No. 5,6 colonies selected by colony PCR at August 11th. | ||
#Prepared 5 ml LBA into glass tubes. | #Prepared 5 ml LBA into glass tubes. | ||
#Re-suspended 2 colony mixture (No. 2 and No. 5 respectively). | #Re-suspended 2 colony mixture (No. 2 and No. 5 respectively). | ||
#Incubated at 37C for 18 hrs. | #Incubated at 37C for 18 hrs. | ||
| - | |||
| - | ==Digestion of RBS-phaB-pSB1A2 as Vector and RBS-eYFP-dT as Insert== | + | ====Digestion of RBS-phaB-pSB1A2 as Vector and RBS-eYFP-dT as Insert==== |
| - | + | ||
To make a construct of RBS-phaB-RBS-eYFP-dT-pSB1A2, we digested RBS-phaB-pSB1A2 with SpeI & PstI, RBS-eYFP-dT with XbaI & PstI. | To make a construct of RBS-phaB-RBS-eYFP-dT-pSB1A2, we digested RBS-phaB-pSB1A2 with SpeI & PstI, RBS-eYFP-dT with XbaI & PstI. | ||
| - | <br /> | + | <br /> |
| - | + | ||
Insert (RBS-eYFP-dT) | Insert (RBS-eYFP-dT) | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 327: | Line 306: | ||
|30 ul | |30 ul | ||
|} | |} | ||
| - | |||
| - | |||
| Line 372: | Line 349: | ||
|HOLD | |HOLD | ||
|} | |} | ||
| - | |||
[[image:HokkaidoU2012 120912 Digestion Insert(RBS-eYFP-dT)-Vector(RBS-phaB-pSB1A2)-Control(pBAD-RBS-eCFP-RBS-pSB1A2).jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120912 Digestion Insert(RBS-eYFP-dT)-Vector(RBS-phaB-pSB1A2)-Control(pBAD-RBS-eCFP-RBS-pSB1A2).jpg|thumb|digestion result]] | ||
| - | |||
| - | ==Ethanol precipitation of ptet-RBS-eYFP-dT-pSB1A2 and pSTV28== | + | ====Ethanol precipitation of ptet-RBS-eYFP-dT-pSB1A2 and pSTV28==== |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
#Centrifuged at 15000 rpm, 15 min at 4C. | #Centrifuged at 15000 rpm, 15 min at 4C. | ||
| Line 384: | Line 358: | ||
#Centrifuged at 15000 rpm, 10 min at 4C. | #Centrifuged at 15000 rpm, 10 min at 4C. | ||
#Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| - | |||
[[image:HokkaidoU2012 120912 Ethapre 20min Insert-Vector.jpg|thumb|ethanol precipitation result]] | [[image:HokkaidoU2012 120912 Ethapre 20min Insert-Vector.jpg|thumb|ethanol precipitation result]] | ||
We confirmed that the concentration of Insert DNA solution is 40 ng/ul and Vector DNA solution is 30 ng/ul. | We confirmed that the concentration of Insert DNA solution is 40 ng/ul and Vector DNA solution is 30 ng/ul. | ||
| - | |||
| - | ==Ligation of pBAD-RBS-eCFP-RBS, Ag43-dT and pSTV28== | + | ====Ligation of pBAD-RBS-eCFP-RBS, Ag43-dT and pSTV28==== |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 406: | Line 377: | ||
|12 ul | |12 ul | ||
|} | |} | ||
| - | |||
Ligation reaction time was in detail below. | Ligation reaction time was in detail below. | ||
| - | |||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 425: | Line 394: | ||
|} | |} | ||
| - | </ | + | <br style="line-height: 0; clear: both;" /> |
| - | + | ||
| + | </div></div> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 13th== | + | ===September 13th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Transformation of RBS-phaB-RBS-eYFP-dT-pSB1A2== | + | ====Transformation of RBS-phaB-RBS-eYFP-dT-pSB1A2==== |
| - | + | ||
Transformation of ligation product into JM109. | Transformation of ligation product into JM109. | ||
#Mixed 2 ul ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul ligation product to 50 ul of thawed competent cells on ice. | ||
| Line 442: | Line 410: | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
#Incubated the plates at 37C for 14 hrs. | #Incubated the plates at 37C for 14 hrs. | ||
| - | |||
| - | ==Aggregation Check of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28== | + | ====Aggregation Check of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28==== |
| - | + | ||
#Mixed 25 ul Arabinose solution (20 %) to 5 ml LBC in glass tube. | #Mixed 25 ul Arabinose solution (20 %) to 5 ml LBC in glass tube. | ||
#Incubated for 24 hrs at 37C. | #Incubated for 24 hrs at 37C. | ||
| - | |||
Result: The construct has aggregated not forming large cluster but small dot cluster, and the supernatant has muddiness. | Result: The construct has aggregated not forming large cluster but small dot cluster, and the supernatant has muddiness. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ====Colony PCR of RBS-phaB-RBS-eYFP-dT-pSB1A2==== | ||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
| Line 518: | Line 480: | ||
We noticed the ligated DNA contains phaB-something-SP sequence and the length is about 1600~1800bp, at least over 1000 bp. We selected No. 1,2 for incubation for plasmid extraction and No. 3,4 for stock at 4C. | We noticed the ligated DNA contains phaB-something-SP sequence and the length is about 1600~1800bp, at least over 1000 bp. We selected No. 1,2 for incubation for plasmid extraction and No. 3,4 for stock at 4C. | ||
| - | |||
| - | + | ====Incubation of RBS-phaB-RBS-eYFP-dT-pSB1A2 for plasmid extraction==== | |
| - | ==Incubation of RBS-phaB-RBS-eYFP-dT-pSB1A2 for plasmid extraction== | + | |
| - | + | ||
#Prepared 2 ml LBC into culture tubes. | #Prepared 2 ml LBC into culture tubes. | ||
#Re-suspended 2 colony mixture (No. 1 and No. 2 respectively). | #Re-suspended 2 colony mixture (No. 1 and No. 2 respectively). | ||
#Incubated at 37C for 14 hrs. | #Incubated at 37C for 14 hrs. | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 14th== | + | ===September 14th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2== | + | ====Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2==== |
| - | + | ||
To make a construct of RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBS-phaA with SpeI and PstI, RBS-phaB-RBS-eYFP-pSB1A2 with XbaI and PstI. | To make a construct of RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBS-phaA with SpeI and PstI, RBS-phaB-RBS-eYFP-pSB1A2 with XbaI and PstI. | ||
<br /><br /> | <br /><br /> | ||
| Line 626: | Line 585: | ||
We suppose the Insert DNA (RBS-phaB-RBS-eYFP-dT) has contained Vector-dimer DNA as contamination. But desired DNA fragment (about 1600 bp) bond also appeared in the result of migration. We picked up the fragment and extracted from gel. | We suppose the Insert DNA (RBS-phaB-RBS-eYFP-dT) has contained Vector-dimer DNA as contamination. But desired DNA fragment (about 1600 bp) bond also appeared in the result of migration. We picked up the fragment and extracted from gel. | ||
| - | |||
| - | ==Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | ====Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
#Centrifuged in 14000 rpm, 30 min at 4C. | #Centrifuged in 14000 rpm, 30 min at 4C. | ||
| Line 635: | Line 592: | ||
#Centrifuged in 15000 rpm, 15 min at 4C. | #Centrifuged in 15000 rpm, 15 min at 4C. | ||
#Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| - | |||
[[image:HokkaidoU2012 120914 Ethapre Insert Vector.jpg|thumb|ethanol precipitation result]] | [[image:HokkaidoU2012 120914 Ethapre Insert Vector.jpg|thumb|ethanol precipitation result]] | ||
We confirmed that the concentration of Insert DNA solution is 20 ng/ul and Vector DNA solution is about 50~60 ng/ul. | We confirmed that the concentration of Insert DNA solution is 20 ng/ul and Vector DNA solution is about 50~60 ng/ul. | ||
| - | |||
| - | ==Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | ====Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 679: | Line 633: | ||
|} | |} | ||
| - | |||
| - | ==Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2== | + | ====Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2==== |
| - | + | ||
Transformation ligation product into JM109. | Transformation ligation product into JM109. | ||
#Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | ||
| Line 693: | Line 645: | ||
We succeeded to transform the cells but noticed that we have used wrong DNA solution. We'll try this DNA synthesis again tomorrow. | We succeeded to transform the cells but noticed that we have used wrong DNA solution. We'll try this DNA synthesis again tomorrow. | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===September 15th=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2==== | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
To make a construct of RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBS-phaA with SpeI and PstI, RBS-phaB-RBS-eYFP-pSB1A2 with XbaI and PstI. | To make a construct of RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBS-phaA with SpeI and PstI, RBS-phaB-RBS-eYFP-pSB1A2 with XbaI and PstI. | ||
<br /><br /> | <br /><br /> | ||
| Line 748: | Line 700: | ||
|20 ul | |20 ul | ||
|} | |} | ||
| - | |||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 768: | Line 719: | ||
|HOLD | |HOLD | ||
|} | |} | ||
| - | |||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 788: | Line 738: | ||
|HOLD | |HOLD | ||
|} | |} | ||
| - | |||
[[image:HokkaidoU2012 120915 Digestion Insert-Vector.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120915 Digestion Insert-Vector.jpg|thumb|digestion result]] | ||
| - | + | ====Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== | |
| - | ==Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
#Centrifuged in 14000 rpm, 30 min at 4C. | #Centrifuged in 14000 rpm, 30 min at 4C. | ||
| Line 800: | Line 747: | ||
#Centrifuged in 15000 rpm, 15 min at 4C. | #Centrifuged in 15000 rpm, 15 min at 4C. | ||
#Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| - | |||
[[image:HokkaidoU2012 120915 Ethapre Insert-Vector.jpg|thumb|ethanol precipitation result]] | [[image:HokkaidoU2012 120915 Ethapre Insert-Vector.jpg|thumb|ethanol precipitation result]] | ||
We confirmed that the concentration of Insert DNA solution is 20 ng/ul and Vector DNA solution is about 50~60 ng/ul. | We confirmed that the concentration of Insert DNA solution is 20 ng/ul and Vector DNA solution is about 50~60 ng/ul. | ||
| - | |||
| - | ==Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | ====Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 825: | Line 769: | ||
|20 ul | |20 ul | ||
|} | |} | ||
| - | |||
Ligation reaction time was in detail below. | Ligation reaction time was in detail below. | ||
| - | |||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 844: | Line 786: | ||
|} | |} | ||
| - | + | ====Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2==== | |
| - | + | ||
| - | ==Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2== | + | |
| - | + | ||
Transformation of ligation product into JM109. | Transformation of ligation product into JM109. | ||
#Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | ||
| Line 856: | Line 795: | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
#Incubated the plates at 37C for 17 hrs. | #Incubated the plates at 37C for 17 hrs. | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 16th== | + | ===September 16th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Digestion of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 and pT7-RBS-pSB1C3== | + | ====Digestion of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 and pT7-RBS-pSB1C3==== |
| - | + | ||
To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3, we digested pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 with EcoRI and SpeI, and pT7-RBS-pSB1C3 with EcoRI and SpeI. As a control, we digested pT7-RBS-pSB1C3 with only EcoRI to confirm the digestibility of This DNA (The digestibility with SpeI has confirmed in August 26th). | To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3, we digested pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 with EcoRI and SpeI, and pT7-RBS-pSB1C3 with EcoRI and SpeI. As a control, we digested pT7-RBS-pSB1C3 with only EcoRI to confirm the digestibility of This DNA (The digestibility with SpeI has confirmed in August 26th). | ||
| - | + | <br /> | |
Insert (pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28) | Insert (pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28) | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 957: | Line 895: | ||
We confirmed 4 bonds in the result of Inset DNA digestion. A Bond which shows the highest concentration and places about 8k ~ 10k bp area would be pBAD-RBS-eCFP-RBS-Ag43-pSTV28 construct. We considered about it bond whether remained from digestion, or at first formed dimer and separated by digestion. In either case, our target product has about 5200 bp and there is appropriate bond. We extracted the target bond from TBE gel and went next step. | We confirmed 4 bonds in the result of Inset DNA digestion. A Bond which shows the highest concentration and places about 8k ~ 10k bp area would be pBAD-RBS-eCFP-RBS-Ag43-pSTV28 construct. We considered about it bond whether remained from digestion, or at first formed dimer and separated by digestion. In either case, our target product has about 5200 bp and there is appropriate bond. We extracted the target bond from TBE gel and went next step. | ||
| - | + | ||
| - | ==Ethanol precipitation of pBAD-RBS-eCFP-RBS-Ag43-dT on pSB1C3== | + | ====Ethanol precipitation of pBAD-RBS-eCFP-RBS-Ag43-dT on pSB1C3==== |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
#Centrifuged in 15000 rpm, 15 min at 4C. | #Centrifuged in 15000 rpm, 15 min at 4C. | ||
| Line 965: | Line 902: | ||
#Centrifuged in 15000 rpm, 10 min at 4C. | #Centrifuged in 15000 rpm, 10 min at 4C. | ||
#Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| - | |||
[[image:HokkaidoU2012 120916 Ethapre Insert Vector.jpg|thumb|ethanol precipitation result]] | [[image:HokkaidoU2012 120916 Ethapre Insert Vector.jpg|thumb|ethanol precipitation result]] | ||
| - | + | ====Ligation of pBAD-RBS-eCFP-RBS-Ag43-dT and pSB1C3==== | |
| - | + | ||
| - | ==Ligation of pBAD-RBS-eCFP-RBS-Ag43-dT and pSB1C3== | + | |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 987: | Line 920: | ||
|10 ul | |10 ul | ||
|} | |} | ||
| - | |||
Ligation reaction time was in detail below. | Ligation reaction time was in detail below. | ||
| - | |||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 1,006: | Line 937: | ||
|} | |} | ||
| - | + | ====Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3==== | |
| - | + | ||
| - | ==Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3== | + | |
| - | + | ||
Transformation ligation product into DH5α. | Transformation ligation product into DH5α. | ||
#Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3 ligation product to 50 ul of thawed competent cells on ice. | ||
| Line 1,019: | Line 947: | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
#Incubated the plates at 37C for 13 hrs. | #Incubated the plates at 37C for 13 hrs. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ====Colony PCR of RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2==== | ||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
| Line 1,077: | Line 1,002: | ||
|} | |} | ||
Cycle:2~4 x 35 | Cycle:2~4 x 35 | ||
| - | |||
We used N1 (DW only) and N2(RBS-phaC-RBS-phaA-RBS-phaB-dT-pSB1A2)as controls. | We used N1 (DW only) and N2(RBS-phaC-RBS-phaA-RBS-phaB-dT-pSB1A2)as controls. | ||
| Line 1,083: | Line 1,007: | ||
[[image:HokkaidoU2012 120916 coloP RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2.jpg|thumb|Colony PCR result]] | [[image:HokkaidoU2012 120916 coloP RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2.jpg|thumb|Colony PCR result]] | ||
| - | |||
We noticed the ligated DNA contains phaB-something-SP sequence and the length is about 1600~1800bp, at least over 1000 bp. We selected No. 1,2 for incubation for plasmid extraction and No. 3,4 for stock at 4C. | We noticed the ligated DNA contains phaB-something-SP sequence and the length is about 1600~1800bp, at least over 1000 bp. We selected No. 1,2 for incubation for plasmid extraction and No. 3,4 for stock at 4C. | ||
| - | |||
| - | ==Incubation of RBS-phaB-RBS-eYFP-dT-pSB1A2 for plasmid extraction== | + | ====Incubation of RBS-phaB-RBS-eYFP-dT-pSB1A2 for plasmid extraction==== |
| - | + | ||
#Prepared 2 ml LBA into culture tubes. | #Prepared 2 ml LBA into culture tubes. | ||
#Re-suspended 2 colony mixture (No. 1 and No. 2 respectively). | #Re-suspended 2 colony mixture (No. 1 and No. 2 respectively). | ||
#Incubated at 37C for 20 hrs. | #Incubated at 37C for 20 hrs. | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | |||
</div></div> | </div></div> | ||
| + | |||
<!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | ||
Latest revision as of 03:21, 27 September 2012
September 10th
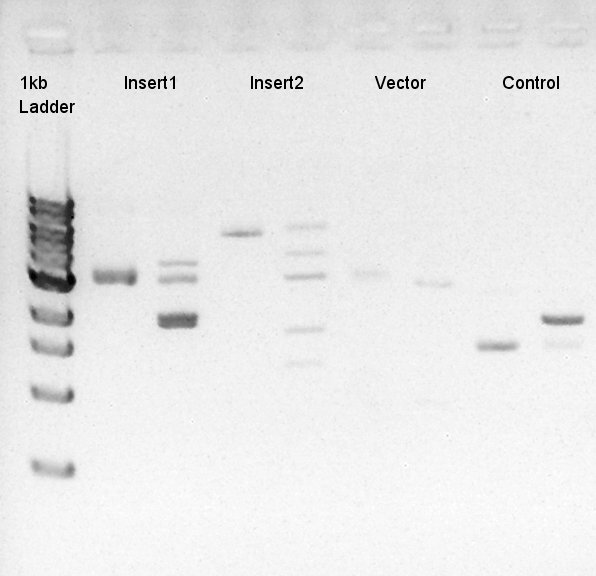
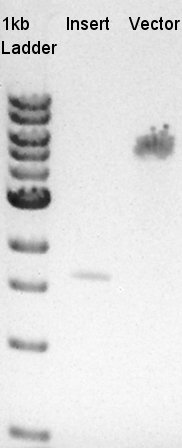
Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2
To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 by 3piece ligation, we digested pBAD-RBS-eCFP-RBS-pSB1A2 by EcoRI and SpeI, Ag43-dT-pSB1AK3 (previously digested by HindIII) by XbaI and NotI, and pSTV28 by EcoRI and NotI. Then we digested pT7-RBS-pSB1C3 by XbaI and SpeI as a control for confirmation of the ability of restriction enzyme.
Insert1 (pBAD-RBS-eCFP-RBS-pSB1A2)
| DNA solution (35 ng/ul) | 17 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 3 ul |
| DW | 8 ul |
| Total | 30 ul |
Insert2 (Ag43-dT-pSB1AK3)
| DNA solution (35 ng/ul) | 25 ul |
| XbaI | 1 ul |
| NotI | 1 ul |
| 10xK buffer | 1.5 ul |
| 100xBSA | 0.3ul |
| DW | 1.5 ul |
| Total | 30 ul |
Vector(pSTV28)
| DNA solution (15 ng/ul) | 9 ul |
| EcoRI | 1 ul |
| NotI | 1 ul |
| 10xH buffer | 2 ul |
| 10xBSA | 0.2 ul |
| DW | 7 ul |
| Total | 20 ul |
control (pT7-RBS-pSB1C3)
| DNA solution (30~40 ng/ul) | 10 ul |
| XbaI | 1 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 6 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 70 | 20 |
| 3 | 4 | HOLD |
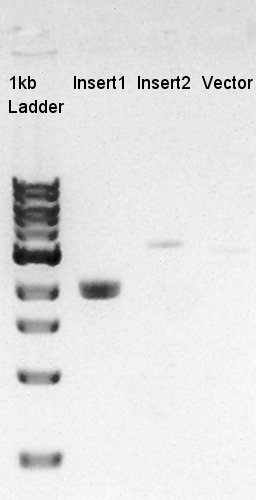
Ethanol precipitation of ptet-RBS-eYFP-dT-pSB1A2 and pSTV28
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 14000 rpm, 30 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We confirmed that the concentration of Insert1 DNA solution is 50 ng/ul, Insert2 DNA solution is 10~15 ng/ul and Vector DNA solution is 10 ng/ul.
Ligation of pBAD-RBS-eCFP-RBS, Ag43-dT and pSTV28
| Vector DNA (10 ng/ul) | 4 ul |
| Insert1 DNA (50 ng/ul) | 2 ul |
| Insert2 DNA (10~15 ng/ul) | 4 ul |
| Ligation Mighty Mix | 10 ul |
| Total | 20 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28
Transformation ligation product into DH5α.
- Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Incubated for 2 hrs to get the resistance to Chloramphenicol.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBC).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 14 hrs.
September 11th
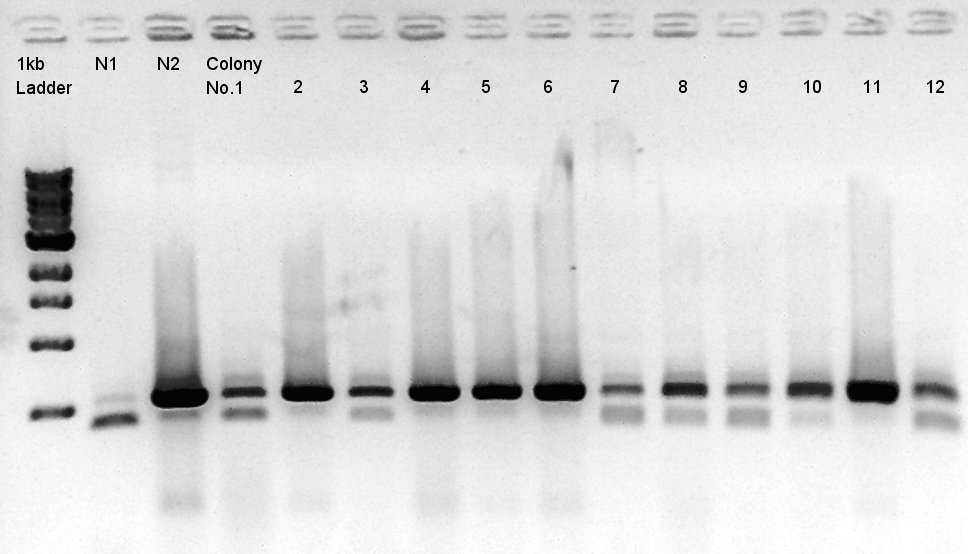
Colony PCR of pBAD-RBS-eCFP-RBS-pSB1A2
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(pbad-f2 primer) | 0.5 ul |
| Reverse Primer(PS-R down primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.3 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
- We noticed that this step was actually 68.9 degree after reaction.
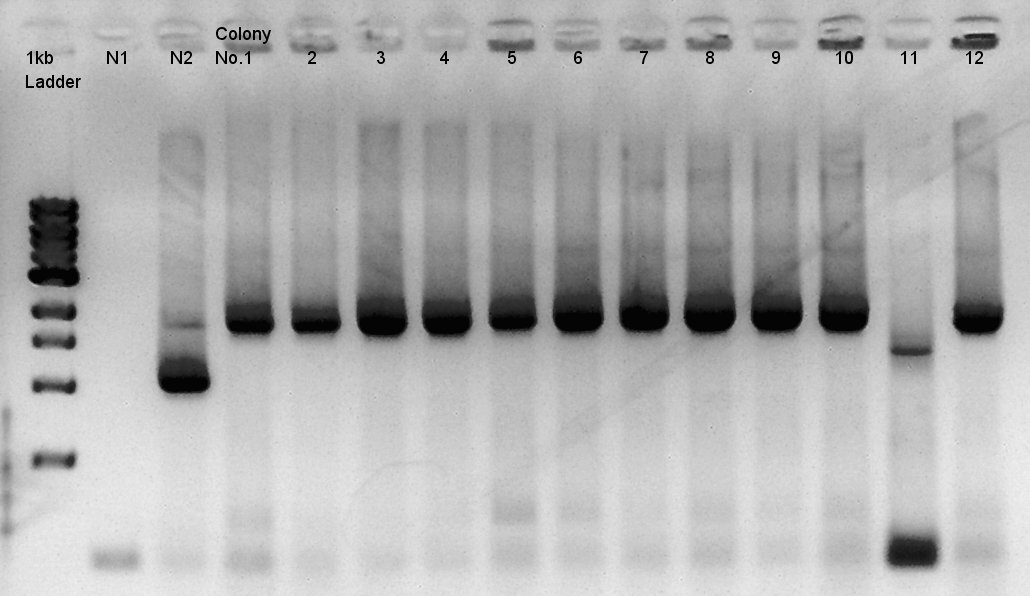
We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls. Desired product is about 542bp.
Because of mis-setting the PCR program, we failed to amplify desired DNA fragment so we retried the colony PCR in same reagent, correct PCR machine program.
We noticed that the ligated DNA contains at least Ag43-dT-BioBrick_Suffix complex. We selected No. 2,4 for incubation for plasmid extraction and No. 5,6 for Aggregation check.
September 12th
Analysis nucleotide sequence
We analyzed the nucleotide sequence of pBAD-RBS-Ag43-dT on pSB1AK3 and pSB1C3. We used these 6 kinds of primers. 100b up EX-F primer, pBAD-f1 primer, pBAD-f2 primer, Ag43-f1 primer, Ag43-f2 primer, Ag43-f3 primer, Ag43-f4 primer, 200b down PS-R primer
Aggregation check of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28
Aggregation check for No. 5,6 colonies selected by colony PCR at August 11th.
- Prepared 5 ml LBA into glass tubes.
- Re-suspended 2 colony mixture (No. 2 and No. 5 respectively).
- Incubated at 37C for 18 hrs.
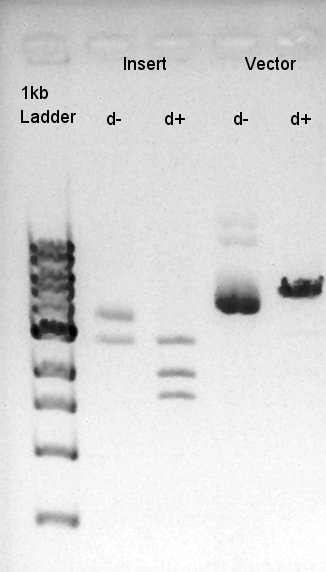
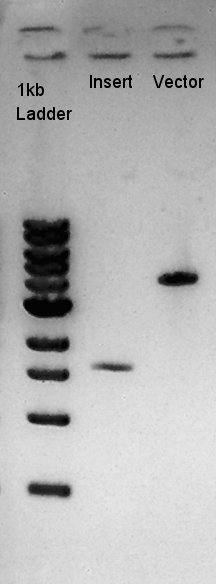
Digestion of RBS-phaB-pSB1A2 as Vector and RBS-eYFP-dT as Insert
To make a construct of RBS-phaB-RBS-eYFP-dT-pSB1A2, we digested RBS-phaB-pSB1A2 with SpeI & PstI, RBS-eYFP-dT with XbaI & PstI.
Insert (RBS-eYFP-dT)
| DNA solution (40 ng/ul) | 25 ul |
| XbaI | 1 ul |
| NotI | 1 ul |
| 10xK buffer | 1.5 ul |
| 100xBSA | 0.3 ul |
| DW | 1.5 ul |
| Total | 30 ul |
Vector(RBS-phaB-pSB1A2)
| DNA solution (35 ng/ul) | 6 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
Ethanol precipitation of ptet-RBS-eYFP-dT-pSB1A2 and pSTV28
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We confirmed that the concentration of Insert DNA solution is 40 ng/ul and Vector DNA solution is 30 ng/ul.
Ligation of pBAD-RBS-eCFP-RBS, Ag43-dT and pSTV28
| Vector DNA (40 ng/ul) | 3 ul |
| Insert DNA (30 ng/ul) | 3 ul |
| Ligation Mighty Mix | 6 ul |
| Total | 12 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
September 13th
Transformation of RBS-phaB-RBS-eYFP-dT-pSB1A2
Transformation of ligation product into JM109.
- Mixed 2 ul ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 14 hrs.
Aggregation Check of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28
- Mixed 25 ul Arabinose solution (20 %) to 5 ml LBC in glass tube.
- Incubated for 24 hrs at 37C.
Result: The construct has aggregated not forming large cluster but small dot cluster, and the supernatant has muddiness.
Colony PCR of RBS-phaB-RBS-eYFP-dT-pSB1A2
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 10 ul |
| Forward Primer(pbad-f2 primer: 10 ng/ul) | 0.8 ul |
| Reverse Primer(PS-R down primer: 10 ng/ul) | 0.8 ul |
| DW | 4.4 ul |
| Total | 20 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 64.4 | 30 |
| 4 | 72 | 180 |
| 5 | 72 | 120 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2(RBS-phaB-pSB1A2)as controls.
Desired product is about 1800~2000bp.
We noticed the ligated DNA contains phaB-something-SP sequence and the length is about 1600~1800bp, at least over 1000 bp. We selected No. 1,2 for incubation for plasmid extraction and No. 3,4 for stock at 4C.
Incubation of RBS-phaB-RBS-eYFP-dT-pSB1A2 for plasmid extraction
- Prepared 2 ml LBC into culture tubes.
- Re-suspended 2 colony mixture (No. 1 and No. 2 respectively).
- Incubated at 37C for 14 hrs.
September 14th
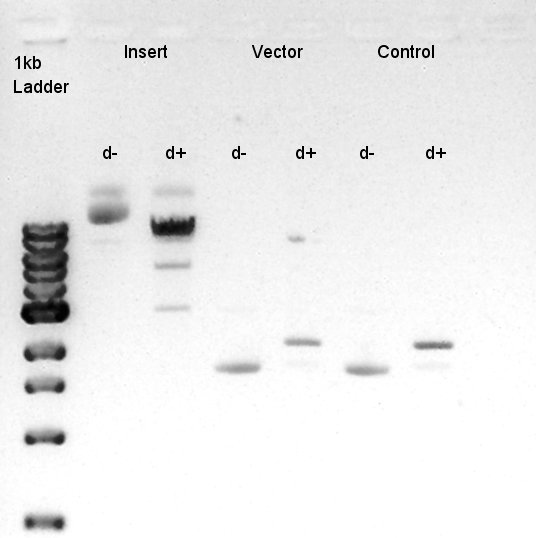
Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2
To make a construct of RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBS-phaA with SpeI and PstI, RBS-phaB-RBS-eYFP-pSB1A2 with XbaI and PstI.
Insert (RBS-phaB-RBS-eYFP-dT)
| DNA solution (30 ng/ul) | 21 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 3 ul |
| DW | 4 ul |
| Total | 30 ul |
Vector(RBS-phaC-RBS-phaA-pSB1A2)
| DNA solution (about 30~40 ng/ul) | 6 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 70 | 20 |
| 3 | 4 | HOLD |
We suppose the Insert DNA (RBS-phaB-RBS-eYFP-dT) has contained Vector-dimer DNA as contamination. But desired DNA fragment (about 1600 bp) bond also appeared in the result of migration. We picked up the fragment and extracted from gel.
Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged in 14000 rpm, 30 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 15 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We confirmed that the concentration of Insert DNA solution is 20 ng/ul and Vector DNA solution is about 50~60 ng/ul.
Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
| Insert DNA (20 ng/ul) | 4 ul |
| Vector DNA (50~60 ng/ul) | 1.5 ul |
| Ligation Mighty Mix | 6 ul |
| DW | 0.5 ul |
| Total | 20 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2
Transformation ligation product into JM109.
- Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 13 hrs.
We succeeded to transform the cells but noticed that we have used wrong DNA solution. We'll try this DNA synthesis again tomorrow.
September 15th
Digestion of eCFP-RBS-pSB1A2 and pBAD-RBS-pSB1A2
To make a construct of RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBS-phaA with SpeI and PstI, RBS-phaB-RBS-eYFP-pSB1A2 with XbaI and PstI.
Insert (RBS-phaB-RBS-eYFP-dT)
| DNA solution (30 ng/ul) | 21 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 3 ul |
| DW | 4 ul |
| Total | 30 ul |
Vector(RBS-phaC-RBS-phaA-pSB1A2)
| DNA solution (about 30~40 ng/ul) | 6 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 70 | 20 |
| 3 | 4 | HOLD |
Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged in 14000 rpm, 30 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 15 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We confirmed that the concentration of Insert DNA solution is 20 ng/ul and Vector DNA solution is about 50~60 ng/ul.
Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
| Insert DNA (20 ng/ul) | 4 ul |
| Vector DNA (50~60 ng/ul) | 1.5 ul |
| Ligation Mighty Mix | 6 ul |
| DW | 0.5 ul |
| Total | 20 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2
Transformation of ligation product into JM109.
- Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 17 hrs.
September 16th
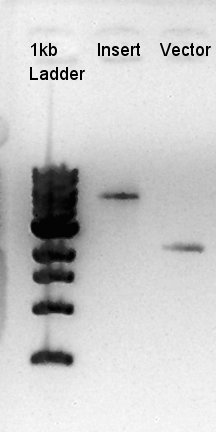
Digestion of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 and pT7-RBS-pSB1C3
To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3, we digested pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 with EcoRI and SpeI, and pT7-RBS-pSB1C3 with EcoRI and SpeI. As a control, we digested pT7-RBS-pSB1C3 with only EcoRI to confirm the digestibility of This DNA (The digestibility with SpeI has confirmed in August 26th).
Insert (pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28)
| DNA solution (about 35ng/ul) | 41 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 5 ul |
| DW | 2 ul |
| Total | 50 ul |
Vector(pT7-RBS-pSB1C3)
| DNA solution (about 30~40 ng/ul) | 2 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 14 ul |
| Total | 20 ul |
control (pT7-RBS-pSB1C3)
| DNA solution (about 30~40 ng/ul) | 2 ul |
| EcoRI | 1 ul |
| 10xH buffer | 1 ul |
| DW | 6 ul |
| Total | 10 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
We confirmed 4 bonds in the result of Inset DNA digestion. A Bond which shows the highest concentration and places about 8k ~ 10k bp area would be pBAD-RBS-eCFP-RBS-Ag43-pSTV28 construct. We considered about it bond whether remained from digestion, or at first formed dimer and separated by digestion. In either case, our target product has about 5200 bp and there is appropriate bond. We extracted the target bond from TBE gel and went next step.
Ethanol precipitation of pBAD-RBS-eCFP-RBS-Ag43-dT on pSB1C3
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged in 15000 rpm, 15 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
Ligation of pBAD-RBS-eCFP-RBS-Ag43-dT and pSB1C3
| Insert DNA | 4 ul |
| Vector DNA | 1 ul |
| Ligation Mighty Mix | 5 ul |
| Total | 10 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3
Transformation ligation product into DH5α.
- Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Incubated for 2 hrs to get the resistance to Chloramphenicol.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBC).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 13 hrs.
Colony PCR of RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 10 ul |
| Forward Primer(phaA-1083bp-F primer: 10 ng/ul) | 0.8 ul |
| Reverse Primer(PS-R down primer: 10 ng/ul) | 0.8 ul |
| DW | 4.4 ul |
| Total | 20 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 68.9 | 30 |
| 4 | 72 | 180 |
| 5 | 72 | 120 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2(RBS-phaC-RBS-phaA-RBS-phaB-dT-pSB1A2)as controls. Desired product is about 1800~2000bp.
We noticed the ligated DNA contains phaB-something-SP sequence and the length is about 1600~1800bp, at least over 1000 bp. We selected No. 1,2 for incubation for plasmid extraction and No. 3,4 for stock at 4C.
Incubation of RBS-phaB-RBS-eYFP-dT-pSB1A2 for plasmid extraction
- Prepared 2 ml LBA into culture tubes.
- Re-suspended 2 colony mixture (No. 1 and No. 2 respectively).
- Incubated at 37C for 20 hrs.
 "
"