Team:LMU-Munich/Weekly Journal
From 2012.igem.org
| (43 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo9.jpg|6}} | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo9.jpg|6}} | ||
| - | [[File: | + | [[File:Weekly journal banner.resized WORDS.JPG|620px|link=]] |
| - | |||
| - | |||
<p></p> | <p></p> | ||
| Line 20: | Line 18: | ||
|[[Team:LMU-Munich/Weekly Journal#July|July]] | |[[Team:LMU-Munich/Weekly Journal#July|July]] | ||
|[[Team:LMU-Munich/Weekly Journal#October|October]] | |[[Team:LMU-Munich/Weekly Journal#October|October]] | ||
| + | |- | ||
| + | |[[Team:LMU-Munich/Weekly Journal# | ]] | ||
| + | |[[Team:LMU-Munich/Weekly Journal# | ]] | ||
| + | |[[Team:LMU-Munich/Weekly Journal#November|November]] | ||
|} | |} | ||
</div> | </div> | ||
| Line 80: | Line 82: | ||
--> | --> | ||
| + | <div class="box" style= "background-color:#e4f1d7"> | ||
| + | ===November=== | ||
| + | </div> | ||
| + | |||
| + | <div class="box"> | ||
| + | '''4-5 November 2012''' | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> Boston Jamboree!!</p> | ||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> OMG! We were finalist! We won "best wiki" together with Slovenia and best "new application"!</p> | ||
| + | </div> | ||
| + | <div class="box" style= "background-color:#e4f1d7"> | ||
| + | ===October=== | ||
| + | </div> | ||
| + | |||
| + | <div class="box"> | ||
| + | '''29 October - 3 November 2012''' | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> Flying across the Atlantic! On our way to the World Championship Jamboree in Boston!</p> | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | '''22-26 October 2012''' | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> Our dedicated presentation and website mini-team (Franzi, Korinna and Tamara) is working hard.</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> Round two of time-lapse microscopy. We decided to try a new media combination and flow speed to see if we can prevent the cell lysis we experienced last week.</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> We brain-stormed some ways to create a device for our '''Sporo'''beads. We asked our Gold Sponsor [https://2012.igem.org/Team:LMU-Munich/Sponsors Semadeni] if they would donate a few 0,20um filter spin columns to use for our envisioned sporo-device. We also batch-produced spores of our strain B53. Even with its modifications, it has definitely not lost its sporulation ability!</p> | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | '''15-19 October 2012''' | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> We got some great ideas from other teams at the European Jamboree about how to better present our project. We're working on a new version of our presentation for Boston.</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> One of our collaborators, the [https://2012.igem.org/Team:Bielefeld-Germany Bielefeld iGEM team], graciously sent us two laccases to use on our '''Sporo'''beads.</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> We wanted to demonstrate the formation of GFP-expressing spores in our cells, so we decided to try time-lapse microscopy. Our first round of time-lapse failed, as the cells spontaneously lysed after 10 hours. We were baffled by this reaction. Well, time to do some research.</p> | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | '''7-12 October 2012''' | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> On Sunday at the Jamboree, we got the great news that our team will take the '''Bead'''zillus project to Boston!</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> Back home to Munich, and a week of much-needed rest for most of the team before heading back to the lab.</p> | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | '''1-6 October 2012''' | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> Working on the final touches to our poster and presentation.</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> The whole team spent the entire Wednesday mixing, rolling, punching, baking and frosting between 600 and 800 '''Bead'''zillus cookies to bring to the European Jamboree poster session!</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> By planes and trains, we made our way to the European Jamboree in the lovely city of Amsterdam.</p> | ||
| + | |||
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> Friday and Saturday were the Jamboree: we gave our presentation, presented our poster, and distributed lots of '''Bead'''zillus cookies! We also met several potential collaboration partners, including [http://www.bsse.ethz.ch/bpl/people/panke Sven Panke] of [https://2012.igem.org/Team:ETH_Zurich ETH Zurich] who proposed the very interesting idea of using our '''Sporo'''beads for protein evolution.</p> | ||
| + | </div> | ||
<div class="box" style= "background-color:#e4f1d7"> | <div class="box" style= "background-color:#e4f1d7"> | ||
===September=== | ===September=== | ||
| Line 87: | Line 144: | ||
'''24-28 September 2012''' | '''24-28 September 2012''' | ||
| - | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> Now, the [[Team:LMU-Munich/Bacillus_BioBricks/Sporovector|'''Sporo''' vector]] is cloned into [http://partsregistry.org/Part:BBa_K823022 pSB<sub>Bs</sub>4S].</p> | + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> Now, the [[Team:LMU-Munich/Bacillus_BioBricks/Sporovector|'''Sporo''' vector]] is cloned into [http://partsregistry.org/Part:BBa_K823022 pSB<sub>Bs</sub>4S] and we are trying to insert [http://partsregistry.org/Part:BBa_K823019 lacZ].</p> |
| - | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> We all are | + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> We all are busy setting up our website and finding the best ways to display our results!</p> |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | Sitting on the computer every day and analyzing the sheer number of microscopy pictures...</p> | + | Sitting on the computer every day and analyzing the sheer number of microscopy pictures... Check out our [https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore data]</p> |
</div> | </div> | ||
| Line 108: | Line 165: | ||
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | [https://2012.igem.org/Team:LMU-Munich/Data/Vectors#pSBBs0K-Pspac Evaluation] of [http://partsregistry.org/Part:BBa_K823026 pSB<sub>''Bs''</sub>0K-P<sub><i>spac</i></sub>] with [http://partsregistry.org/Part:BBa_K823019 ''lacZ'']</p> | + | [https://2012.igem.org/Team:LMU-Munich/Data/Vectors#pSBBs0K-Pspac Evaluation] of [http://partsregistry.org/Part:BBa_K823026 pSB<sub>''Bs''</sub>0K-P<sub><i>spac</i></sub>] with [http://partsregistry.org/Part:BBa_K823019 ''lacZ''].</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/0/0f/LMU-Munich-Invertersign.png" height=30"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/0/0f/LMU-Munich-Invertersign.png" height=30"/></a></html> | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] measured | + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] measured quantitively via [https://2012.igem.org/File:LMU_Inverter_graph.png β-Galactosidase assay].</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | For our '''Suicide''' switch, the first plate reader measurement of ''Bacillus subtilis'' strain W168 containing ''thrC''::P<sub>''spoIVB''</sub>-''ecf41<sub>Bli aa 1-204</sub>'' (through transformation with pSBBs4S-P<sub>''spoIVB''</sub>-''ecf41<sub>Bli aa 1-204</sub>'') and ''sacA''::P<sub>''ydfG''</sub>-''luxABCDE'' (through transformation with pSBBs3C-''luxABCDE''-P<sub>''ydfG''</sub>) was performed.</p> | |
| + | |||
| + | <p align="justify"><html><a> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| + | Knockouts: One last run of the germination assay on our triple and quadruple mutants. All results of these assays can be found compiled on our [https://2012.igem.org/Team:LMU-Munich/Data/Knockout data page].</p> | ||
</div> | </div> | ||
| Line 124: | Line 185: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | To clean up our '''Sporo'''beads from vegetative ''B. subtilis'' cells we tried three different methods: French Press, sonification and lysozymes. A great difference was observed after the [https://2012.igem.org/Team:LMU-Munich/Data/Sporepurification treatment with lysozyme]! Furthermore the lysozyme did not damage our fusion protein as GFP fluorescence was still obtained in microscopy!</p> | + | To clean up our '''Sporo'''beads from vegetative ''B. subtilis'' cells we tried three different methods: French Press, sonification and lysozymes. A great difference was observed after the [https://2012.igem.org/Team:LMU-Munich/Data/Sporepurification treatment with lysozyme]! Furthermore, the lysozyme did not damage our fusion protein, as GFP fluorescence was still obtained in microscopy!</p> |
| + | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| + | P<sub>''lepA''</sub> was fused into the finished vector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823025 pSB<sub>''BS''</sub>3C-luxABCDE] to evaluate this BioBrick vector and compare in to the version where there is still one forbidden restriction site in it.</p> | ||
| + | |||
| + | <p align="justify"><html><a> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| + | For the '''Suicide''' switch, P<sub>''spoIVB''</sub>-''ecf41<sub>Bli aa 1-204</sub>'' in pSB<sub>BS</sub>4S, and P<sub>''ydfG''</sub>/P<sub>''sspK''</sub>/P<sub>''spoIVB''</sub> in pSB<sub>BS</sub>3C-''luxABCDE'' were brought into ''B. subtilis''.</p> | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | More work on the knockouts: We were really surprised at our great results so far, and repeated the germination assay on our triple and quadruple mutants once again.</p> | |
</div> | </div> | ||
| Line 136: | Line 203: | ||
<p align="justify"><html><a "> | <p align="justify"><html><a "> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
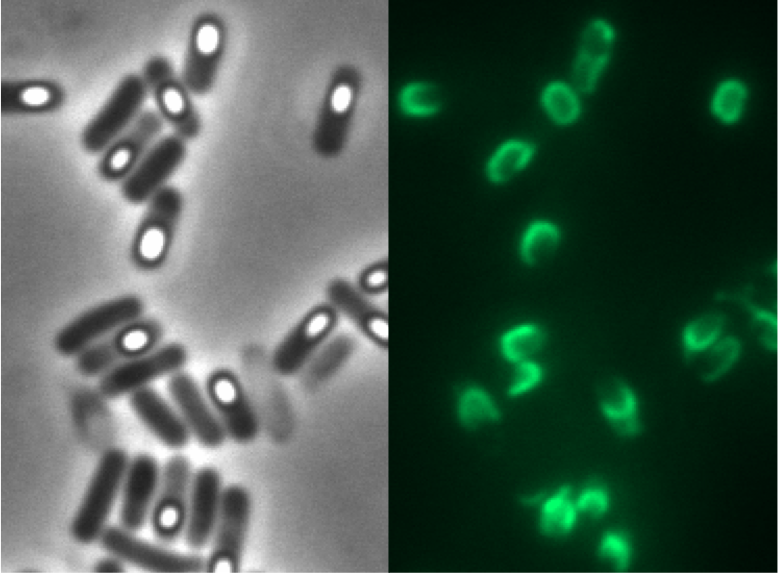
| - | All our CotZ-GFP variants were examined with fluorescence microscopy! The brightest spores derived from our P<sub>''cotyz''</sub>-''cotZ''rep-''gfp''-terminator | + | All of our CotZ-GFP variants were examined with fluorescence microscopy! The brightest spores derived from our B 53 strain (containing P<sub>''cotyz''</sub>-''cotZ''rep-''gfp''-terminator).</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | Plates of our spores diluted at 10<sup>-2</sup>, 10<sup>-4</sup> and 10<sup>-6</sup> | + | We worked more on the knockouts aspect of the '''Germination'''STOP module. We ran another germination assay for our triple and quadruple mutants. Plates of our spores diluted at 10<sup>-2</sup>, 10<sup>-4</sup> and 10<sup>-6</sup> show NO GERMINATION for our mutants, and plenty of germination for the WT168 positive control! We will try plating undiluted mutant spores to see if any germination occurs at higher concentrations.</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823042 | + | More work on the '''Suicide''' switch! [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823042 P<sub>''sspK''</sub>], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823048 P<sub>''spoIVB''</sub>] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823041 P<sub>''ydfG''</sub>] were cloned into pSB1C3 and verified by sequencing. <br> P<sub>''spoIVB''</sub>-''ecf41<sub>Bli aa1-204</sub>'' were cloned into pSB<sub>BS</sub>4S and verified by sequencing. <br> P<sub>''ydfG''</sub>/P<sub>''sspK''</sub>/P<sub>''spoIVB''</sub> were brought into pSB<sub>BS</sub>3C-''luxABCDE''.</p> |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | [http://partsregistry.org/Part:BBa_K823019 lacZ] was succesfully cloned into [http://partsregistry.org/Part:BBa_K823024 pSB<sub>''Bs''</sub>4S-P<sub><i>Xyl</i></sub>] and is functional (blue colonies with IPTG and X-Gal)</p> | + | [http://partsregistry.org/Part:BBa_K823019 lacZ] was succesfully cloned into [http://partsregistry.org/Part:BBa_K823024 pSB<sub>''Bs''</sub>4S-P<sub><i>Xyl</i></sub>] and is functional (blue colonies with IPTG and X-Gal).</p> |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| Line 163: | Line 230: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | Finally, we got our first glowing spores!! After 4 months of hard work we have the first proof that this module works.</p> | + | Finally, we got our first glowing spores!! After 4 months of hard work, we have the first proof that this module works.</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | More on germination knockouts -- we created quadruple mutants using two variations on past mutants: ''cwlD''::kan + ''cwlJ''::spec + ''gerD''::cm + ''sleB''::mls and ''gerD''::cm + ''sleB''::mls + ''cwlJ''::spec + ''cwlD''::kan. See our [https://2012.igem.org/Team:LMU-Munich/Strains Strains Collection.] | |
Germination assay was performed on triple and quadruple mutants.</p> | Germination assay was performed on triple and quadruple mutants.</p> | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823044 MazF] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823043 | + | For the '''Suicide''' switch, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823044 MazF] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823043 Ecf41<sub>Bli aa 1-204</sub>] were cloned into pSB1C3 and verified by sequencing.</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | The BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 lacZ] for ''B. subtilis'' was shown to be functional in <b>pSB<sub>Bs</sub>0K-P<sub><i>spac</i></sub> </b> in ''E. coli'' and ''B. subtilis'' | + | The BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 lacZ] for ''B. subtilis'' was shown to be functional in <b>pSB<sub>Bs</sub>0K-P<sub><i>spac</i></sub> </b> in ''E. coli'' and ''B. subtilis'' (blue color on plates with IPTG and X-Gal).</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | The genes [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 luc+] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 mKate2], synthesized by [https://2012.igem.org/Team:LMU-Munich/Sponsors GeneArt] were successfully cloned into pSB1C3 and sequenced.</p> | + | The genes [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 luc+] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 mKate2], synthesized by [https://2012.igem.org/Team:LMU-Munich/Sponsors GeneArt], were successfully cloned into pSB1C3 and sequenced.</p> |
</div> | </div> | ||
| Line 188: | Line 255: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/0/0f/LMU-Munich-Invertersign.png" height=30"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/0/0f/LMU-Munich-Invertersign.png" height=30"/></a></html> | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] with | + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] with lacZα was finished and it works qualitatively.</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | Good and bad news this week. First the good | + | Good and bad news this week. First the good news: Another big step towards the GFP-Sporobeads is done! We have the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823049 CotZ constructs] in pSB<sub>BS</sub>1C! Hopefully it will integrate easily! :D |
| - | Now the bad | + | Now the bad news: Finally we could start with the [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters P<sub>cgeA</sub> evaluation]! But contrary to our expectations, this promoter did not show any activity. </p> |
</div> | </div> | ||
| Line 201: | Line 268: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | For the knockouts, we used last week's double mutants to create triple mutants as follows: ''cwlD''::kan + ''sleB''::mls + ''cwlJ''::spec ; ''cwlD''::kan + ''sleB''::mls + ''gerD''::cm ; ''cwlD''::kan + ''cwlJ''::spec + ''gerD''::cm ; and ''gerD''::cm + ''sleB''::mls + ''cwlJ''::spec. See our [https://2012.igem.org/Team:LMU-Munich/Strains Strains Collection].</p> | |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | ß-glactosidase assay of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103] in <b>pSB<sub>Bs</sub>1C-<i>lacZ</i> </b> in ''B. subtilis''. | + | ß-glactosidase assay of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103] in <b>pSB<sub>Bs</sub>1C-<i>lacZ</i> </b> in ''B. subtilis'' was performed. |
The xylose-inducible promoter with the according repressor (which has a constitutive promoter, RBS and terminator) [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823015 ''xylR''-P<sub><i>Xyl</i></sub>] was cloned into pSB1C3 and sequenced.</p> | The xylose-inducible promoter with the according repressor (which has a constitutive promoter, RBS and terminator) [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823015 ''xylR''-P<sub><i>Xyl</i></sub>] was cloned into pSB1C3 and sequenced.</p> | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | At last Bacillus was keen on integrating the pSB<sub>BS</sub>3C-luxABCDE-P<sub>''cgeA''</sub>!! And the clean deletions of CotZ and CgeA worked out, too!</p> | + | At last ''Bacillus'' was keen on integrating the pSB<sub>BS</sub>3C-luxABCDE-P<sub>''cgeA''</sub>!! And the clean deletions of CotZ and CgeA worked out, too!</p> |
</div> | </div> | ||
| Line 218: | Line 285: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | We created four double-mutants using resistance-cassettes to knock out germination genes as follows: ''cwlD''::kan + ''sleB''::mls ; ''gerD''::cm + ''sleB''::mls ; ''gerD''::cm + ''cwlD''::kan ; ''cwlJ''::spec + ''cwlD''::kan. We also created the resistance cassette knockout ''cwlB''::kan.</p> | + | Germination knockouts: We created four double-mutants using resistance-cassettes to knock out germination genes as follows: ''cwlD''::kan + ''sleB''::mls ; ''gerD''::cm + ''sleB''::mls ; ''gerD''::cm + ''cwlD''::kan ; ''cwlJ''::spec + ''cwlD''::kan. See our [https://2012.igem.org/Team:LMU-Munich/Strains Strains Collection]. Of these double mutants, we tested three strains in our germination assay. We also created the resistance cassette knockout ''cwlB''::kan.</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
| Line 227: | Line 294: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | The final Promoter-CotZ-GFP-Terminator constructs in pSB1C3 are ready!! | + | The final Promoter-CotZ-GFP-Terminator constructs in pSB1C3 are ready!! Our Promoter-CgeAmut constructs are finally finished too.</p> |
</div> | </div> | ||
| Line 248: | Line 315: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> | ||
| - | This week is our Human | + | This week is our Human Practice week! |
| - | + | From the 23<sup>rd</sup> to 25<sup>th</sup> of July we enjoyed the [https://2012.igem.org/Team:LMU-Munich/Human_Practice/CAS_Conference CAS SynBio conference] and the visit of 10 iGEM teams! | |
This weekend, high school students participated in our [https://2012.igem.org/Team:LMU-Munich/Human_Practice/Students_Practical_Course Students Practical Course] for three days and developed their ideas for useful '''Sporo'''beads! | This weekend, high school students participated in our [https://2012.igem.org/Team:LMU-Munich/Human_Practice/Students_Practical_Course Students Practical Course] for three days and developed their ideas for useful '''Sporo'''beads! | ||
It was a great week! </p> | It was a great week! </p> | ||
| Line 255: | Line 322: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | Plate reader measurements of the ''Bacillus'' promoters [http://partsregistry.org/Part:BBa_K823000 P<sub>''liaG''</sub>], [http://partsregistry.org/Part:BBa_K823001 P<sub>''liaI''</sub>] and [http://partsregistry.org/Part:BBa_K823002 P<sub>''lepA''</sub>] finished. Look at [https://2012.igem.org/Team:LMU-Munich/Data Data]!</p> | + | Plate reader measurements of the ''Bacillus'' promoters [http://partsregistry.org/Part:BBa_K823000 P<sub>''liaG''</sub>], [http://partsregistry.org/Part:BBa_K823001 P<sub>''liaI''</sub>] and [http://partsregistry.org/Part:BBa_K823002 P<sub>''lepA''</sub>] are finished. Look at [https://2012.igem.org/Team:LMU-Munich/Data Data]!</p> |
</div> | </div> | ||
| Line 263: | Line 330: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> | ||
| - | Not many of us are working on our modules this week, as we are helping out for [https://2012.igem.org/Team:LMU-Munich/Human_Practice/CAS_Conference CAS SynBio conference] and organising the [https://2012.igem.org/Team:LMU-Munich/Human_Practice/Students_Practical_Course Students Practical Course].</p> | + | Not many of us are working on our modules this week, as we are helping out for the [https://2012.igem.org/Team:LMU-Munich/Human_Practice/CAS_Conference CAS SynBio conference] and organising the [https://2012.igem.org/Team:LMU-Munich/Human_Practice/Students_Practical_Course Students Practical Course].</p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
| Line 278: | Line 345: | ||
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | The vectors [http://partsregistry.org/Part:BBa_K823024 <b>pSB<sub>Bs</sub>4S-P<sub><i>Xyl</i></sub> </b>], [http://partsregistry.org/Part:BBa_K823021 <b>pSB<sub>''Bs''</sub>1C-''lacZ''</b>] and [http://partsregistry.org/Part:BBa_K823022 <b>pSB<sub>Bs</sub>4S </b>] were succesfully completed and tested by restriction digest as well as red colony color. </p> | + | The vectors [http://partsregistry.org/Part:BBa_K823024 <b>pSB<sub>Bs</sub>4S-P<sub><i>Xyl</i></sub> </b>], [http://partsregistry.org/Part:BBa_K823021 <b>pSB<sub>''Bs''</sub>1C-''lacZ''</b>] and [http://partsregistry.org/Part:BBa_K823022 <b>pSB<sub>Bs</sub>4S </b>] were succesfully completed and tested by restriction digest as well as checked for red colony color. </p> |
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | We started with the clean deletions of CgeA and CotZ this week | + | We started with the clean deletions of CgeA and CotZ this week. It seems like it will take forever if you judge from the protocol length... What took even longer is the pSB<sub>BS</sub>3C-luxABCDE-P<sub>''cgeA''</sub>, but it is finished now!</p> |
</div> | </div> | ||
| Line 290: | Line 357: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | In the knockouts part of the '''Germination'''STOP module, clean deletions of germination genes ''sleB'' and ''cwlB'' from PCR were accomplished. DNA was purified and frozen to be later transformed with ''Bacillus''.</p> | |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| Line 301: | Line 368: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | Our first plate reader experiments with [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters P<sub>''cotyz''</sub>] and [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters P<sub>''cotv''</sub>] are running! Like expected they both show activity in the late stationary phase, thus during sporulation!</p> | + | Our first plate reader experiments with [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters P<sub>''cotyz''</sub>] and [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters P<sub>''cotv''</sub>] are running! Like expected, they both show activity in the late stationary phase, thus during sporulation!</p> |
</div> | </div> | ||
| Line 314: | Line 381: | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
pSB<sub>''BS''</sub>3C-luxABCDE-P<sub>''cotyz''</sub> and pSB<sub>''BS''</sub>3C-luxABCDE-P<sub>''cotv''</sub> integrated into the ''B. subtilis'' genome! | pSB<sub>''BS''</sub>3C-luxABCDE-P<sub>''cotyz''</sub> and pSB<sub>''BS''</sub>3C-luxABCDE-P<sub>''cotv''</sub> integrated into the ''B. subtilis'' genome! | ||
| - | Bad luck, we found an additional AgeI site in ''cgeA'' | + | Bad luck, we found an additional AgeI site in ''cgeA''. It seems that this is the reason why we could not fuse ''gfp'' to it... Mutagenesis primers are designed and ordered!</p> |
</div> | </div> | ||
| Line 330: | Line 397: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | Clean deletions of germination genes ''cwlD'', ''cwlJ'', and ''gerD'' from PCR accomplished. DNA purified and frozen to be later transformed with ''Bacillus''.</p> | + | Knockouts: Clean deletions of germination genes ''cwlD'', ''cwlJ'', and ''gerD'' from PCR accomplished. DNA purified and frozen to be later transformed with ''Bacillus''. Edit: the clean deletions were never used for transformation, but remain ready for future use.</p> |
</div> | </div> | ||
| Line 359: | Line 426: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | pSB<sub>''BS''</sub>3C-''luxABCDE''-P<sub>''cotyz''</sub> and pSB<sub>''BS''</sub>3C-luxABCDE-P<sub>''cotv''</sub> are ready for transformation into Bacillus! </p> | + | pSB<sub>''BS''</sub>3C-''luxABCDE''-P<sub>''cotyz''</sub> and pSB<sub>''BS''</sub>3C-luxABCDE-P<sub>''cotv''</sub> are ready for transformation into ''Bacillus''! </p> |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | Promoters [http://partsregistry.org/Part:BBa_K823000 P<sub>''liaG''</sub>], [http://partsregistry.org/Part:BBa_K823001 P<sub>''liaI''</sub>], [http://partsregistry.org/Part:BBa_K823003 P<sub>''veg''</sub>] and [http://partsregistry.org/Part:BBa_K823002 P<sub>''lepA''</sub>] are now in the vector pSB1C3 as BioBrick standard for the registry. </p> | + | Promoters [http://partsregistry.org/Part:BBa_K823000 P<sub>''liaG''</sub>], [http://partsregistry.org/Part:BBa_K823001 P<sub>''liaI''</sub>], [http://partsregistry.org/Part:BBa_K823003 P<sub>''veg''</sub>] and [http://partsregistry.org/Part:BBa_K823002 P<sub>''lepA''</sub>] are now in the vector pSB1C3 as per the BioBrick standard for the registry. </p> |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| Line 377: | Line 444: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | Combining the BioBricks and integrating them into different vectors works | + | Combining the BioBricks and integrating them into different vectors works well! But still there are constructs missing... We'll keep on working! The fusion of the up and down fragment of CotZ finally works!</p> |
</div> | </div> | ||
| Line 385: | Line 452: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=40"/></a></html> | ||
| - | The last weeks we tried to fuse the up and down fragment of our spore crust gene together for its clean deletion. For CgeA we have the first fused fragments! CotZ needs a little more attention :)</p> | + | The last weeks we tried to fuse the up and down fragment of our spore crust gene together for its clean deletion. For CgeA we have the first fused fragments! CotZ needs a little more attention. :)</p> |
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| Line 417: | Line 484: | ||
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | The cloning of the reporter vector [http://partsregistry.org/Part:BBa_K823021 pSB<sub>''Bs''</sub>1C-''lacZ''] | + | The cloning of the reporter vector [http://partsregistry.org/Part:BBa_K823021 pSB<sub>''Bs''</sub>1C-''lacZ''] was finished.</p> |
</div> | </div> | ||
| Line 425: | Line 492: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | For the knockouts, we created the single mutant ''cwlD''::kan.</p> | |
</div> | </div> | ||
| Line 433: | Line 500: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | In our germination genes knockout work, we successfully created our first single knockouts of germination genes using a resistance cassettes: ''sleB''::mls, ''gerD''::cm and ''cwlJ''::spec. See our [https://2012.igem.org/Team:LMU-Munich/Strains Strains Collection]. | |
We tried knocking out ''cwlB'' using the kan resistance cassette. Mutants of ''cwlB''::kan grew very poorly.</p> | We tried knocking out ''cwlB'' using the kan resistance cassette. Mutants of ''cwlB''::kan grew very poorly.</p> | ||
<p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | <p align="justify"><html><a><img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=40"/></a></html> | ||
| - | With [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 <b>pSB<sub>Bs</sub>0K-P<sub><i>spac</i></sub> </b>] our first vector for our <b>B</b>acillus <b>B</b>io<b>B</b>rick <b>B</b>ox was completed.</p> | + | With [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 <b>pSB<sub>Bs</sub>0K-P<sub><i>spac</i></sub> </b>] our first vector for our <b>''B''</b>''acillus'' <b>B</b>io<b>B</b>rick <b>B</b>ox was completed.</p> |
</div> | </div> | ||
| Line 449: | Line 516: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | We tried knocking out ''cwlB'' using the tet resistance cassette. Mutants of ''cwlB''::tet grew very poorly.</p> | + | Knockouts: We tried knocking out ''cwlB'' using the tet resistance cassette. Mutants of ''cwlB''::tet grew very poorly.</p> |
</div> | </div> | ||
| Line 457: | Line 524: | ||
<p align="justify"><html><a> | <p align="justify"><html><a> | ||
<img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=40"/></a></html> | ||
| - | + | For our germination gene knockouts, we decided which genes to knock out for the germination stop. Based on the work of [http://www.ncbi.nlm.nih.gov/pubmed/19554258 J. Kim and W. Schumann (2009)], we decided to knock out genes ''cwlB'', ''gerD'', ''cwlJ'', and ''sleB''. From the research of [http://www.ncbi.nlm.nih.gov/pubmed/11466293 B. Setlow et al (2001)], we also chose ''cwlD''.</p> | |
| + | </div> | ||
| + | |||
| + | <div class="box"> | ||
| + | '''5-9 March 2012''' | ||
| + | |||
| + | <p align="justify"><html><a> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> | ||
| + | We spent two full days (and nights) together planning our project in our "iGEM days." The idea of '''Bead'''zillus was born, and we got to know each other better as we divided the modules of the project, and learned each others' strengths and experience in different fields such as microbiology, ecology and biochemistry.</p> | ||
</div> | </div> | ||
| Line 466: | Line 541: | ||
<div class="box"> | <div class="box"> | ||
'''20-24 February 2012''' | '''20-24 February 2012''' | ||
| - | + | <p align="justify"><html><a> | |
| - | Team fully formed! | + | <img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=25"/></a></html> |
| + | Team fully formed!</p> | ||
</div> | </div> | ||
Latest revision as of 13:37, 14 November 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


|
|
November
4-5 November 2012
 OMG! We were finalist! We won "best wiki" together with Slovenia and best "new application"!
OMG! We were finalist! We won "best wiki" together with Slovenia and best "new application"!
October
29 October - 3 November 2012
 Flying across the Atlantic! On our way to the World Championship Jamboree in Boston!
Flying across the Atlantic! On our way to the World Championship Jamboree in Boston!
22-26 October 2012
 Our dedicated presentation and website mini-team (Franzi, Korinna and Tamara) is working hard.
Our dedicated presentation and website mini-team (Franzi, Korinna and Tamara) is working hard.
 Round two of time-lapse microscopy. We decided to try a new media combination and flow speed to see if we can prevent the cell lysis we experienced last week.
Round two of time-lapse microscopy. We decided to try a new media combination and flow speed to see if we can prevent the cell lysis we experienced last week.
 We brain-stormed some ways to create a device for our Sporobeads. We asked our Gold Sponsor Semadeni if they would donate a few 0,20um filter spin columns to use for our envisioned sporo-device. We also batch-produced spores of our strain B53. Even with its modifications, it has definitely not lost its sporulation ability!
We brain-stormed some ways to create a device for our Sporobeads. We asked our Gold Sponsor Semadeni if they would donate a few 0,20um filter spin columns to use for our envisioned sporo-device. We also batch-produced spores of our strain B53. Even with its modifications, it has definitely not lost its sporulation ability!
15-19 October 2012
 We got some great ideas from other teams at the European Jamboree about how to better present our project. We're working on a new version of our presentation for Boston.
We got some great ideas from other teams at the European Jamboree about how to better present our project. We're working on a new version of our presentation for Boston.
 One of our collaborators, the Bielefeld iGEM team, graciously sent us two laccases to use on our Sporobeads.
One of our collaborators, the Bielefeld iGEM team, graciously sent us two laccases to use on our Sporobeads.
 We wanted to demonstrate the formation of GFP-expressing spores in our cells, so we decided to try time-lapse microscopy. Our first round of time-lapse failed, as the cells spontaneously lysed after 10 hours. We were baffled by this reaction. Well, time to do some research.
We wanted to demonstrate the formation of GFP-expressing spores in our cells, so we decided to try time-lapse microscopy. Our first round of time-lapse failed, as the cells spontaneously lysed after 10 hours. We were baffled by this reaction. Well, time to do some research.
7-12 October 2012
 On Sunday at the Jamboree, we got the great news that our team will take the Beadzillus project to Boston!
On Sunday at the Jamboree, we got the great news that our team will take the Beadzillus project to Boston!
 Back home to Munich, and a week of much-needed rest for most of the team before heading back to the lab.
Back home to Munich, and a week of much-needed rest for most of the team before heading back to the lab.
1-6 October 2012
 Working on the final touches to our poster and presentation.
Working on the final touches to our poster and presentation.
 The whole team spent the entire Wednesday mixing, rolling, punching, baking and frosting between 600 and 800 Beadzillus cookies to bring to the European Jamboree poster session!
The whole team spent the entire Wednesday mixing, rolling, punching, baking and frosting between 600 and 800 Beadzillus cookies to bring to the European Jamboree poster session!
 By planes and trains, we made our way to the European Jamboree in the lovely city of Amsterdam.
By planes and trains, we made our way to the European Jamboree in the lovely city of Amsterdam.
 Friday and Saturday were the Jamboree: we gave our presentation, presented our poster, and distributed lots of Beadzillus cookies! We also met several potential collaboration partners, including [http://www.bsse.ethz.ch/bpl/people/panke Sven Panke] of ETH Zurich who proposed the very interesting idea of using our Sporobeads for protein evolution.
Friday and Saturday were the Jamboree: we gave our presentation, presented our poster, and distributed lots of Beadzillus cookies! We also met several potential collaboration partners, including [http://www.bsse.ethz.ch/bpl/people/panke Sven Panke] of ETH Zurich who proposed the very interesting idea of using our Sporobeads for protein evolution.
September
24-28 September 2012
 Now, the Sporo vector is cloned into [http://partsregistry.org/Part:BBa_K823022 pSBBs4S] and we are trying to insert [http://partsregistry.org/Part:BBa_K823019 lacZ].
Now, the Sporo vector is cloned into [http://partsregistry.org/Part:BBa_K823022 pSBBs4S] and we are trying to insert [http://partsregistry.org/Part:BBa_K823019 lacZ].
 We all are busy setting up our website and finding the best ways to display our results!
We all are busy setting up our website and finding the best ways to display our results!
 Sitting on the computer every day and analyzing the sheer number of microscopy pictures... Check out our data
Sitting on the computer every day and analyzing the sheer number of microscopy pictures... Check out our data
17-21 September 2012
 Last microscopy experiments with the clean deletion mutants. They are glowing even brighter!
Last microscopy experiments with the clean deletion mutants. They are glowing even brighter!
 Finished cloning of [http://partsregistry.org/Part:BBa_K823029 mKate2] constructs. Now the evaluation of this reporter BioBrick with three different promoters, [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823002 PlepA] and the Anderson promoter [http://partsregistry.org/Part:BBa_K823005 J23101], can start.
Finished cloning of [http://partsregistry.org/Part:BBa_K823029 mKate2] constructs. Now the evaluation of this reporter BioBrick with three different promoters, [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823002 PlepA] and the Anderson promoter [http://partsregistry.org/Part:BBa_K823005 J23101], can start.
 Finished cloning of Sporo vector in pSB1C3.
Finished cloning of Sporo vector in pSB1C3.
 Evaluation of [http://partsregistry.org/Part:BBa_K823026 pSBBs0K-Pspac] with [http://partsregistry.org/Part:BBa_K823019 lacZ].
Evaluation of [http://partsregistry.org/Part:BBa_K823026 pSBBs0K-Pspac] with [http://partsregistry.org/Part:BBa_K823019 lacZ].
 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] measured quantitively via β-Galactosidase assay.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] measured quantitively via β-Galactosidase assay.
 For our Suicide switch, the first plate reader measurement of Bacillus subtilis strain W168 containing thrC::PspoIVB-ecf41Bli aa 1-204 (through transformation with pSBBs4S-PspoIVB-ecf41Bli aa 1-204) and sacA::PydfG-luxABCDE (through transformation with pSBBs3C-luxABCDE-PydfG) was performed.
For our Suicide switch, the first plate reader measurement of Bacillus subtilis strain W168 containing thrC::PspoIVB-ecf41Bli aa 1-204 (through transformation with pSBBs4S-PspoIVB-ecf41Bli aa 1-204) and sacA::PydfG-luxABCDE (through transformation with pSBBs3C-luxABCDE-PydfG) was performed.
 Knockouts: One last run of the germination assay on our triple and quadruple mutants. All results of these assays can be found compiled on our data page.
Knockouts: One last run of the germination assay on our triple and quadruple mutants. All results of these assays can be found compiled on our data page.
10-14 September 2012
 To clean up our Sporobeads from vegetative B. subtilis cells we tried three different methods: French Press, sonification and lysozymes. A great difference was observed after the treatment with lysozyme! Furthermore, the lysozyme did not damage our fusion protein, as GFP fluorescence was still obtained in microscopy!
To clean up our Sporobeads from vegetative B. subtilis cells we tried three different methods: French Press, sonification and lysozymes. A great difference was observed after the treatment with lysozyme! Furthermore, the lysozyme did not damage our fusion protein, as GFP fluorescence was still obtained in microscopy!
 PlepA was fused into the finished vector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823025 pSBBS3C-luxABCDE] to evaluate this BioBrick vector and compare in to the version where there is still one forbidden restriction site in it.
PlepA was fused into the finished vector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823025 pSBBS3C-luxABCDE] to evaluate this BioBrick vector and compare in to the version where there is still one forbidden restriction site in it.
 For the Suicide switch, PspoIVB-ecf41Bli aa 1-204 in pSBBS4S, and PydfG/PsspK/PspoIVB in pSBBS3C-luxABCDE were brought into B. subtilis.
For the Suicide switch, PspoIVB-ecf41Bli aa 1-204 in pSBBS4S, and PydfG/PsspK/PspoIVB in pSBBS3C-luxABCDE were brought into B. subtilis.
 More work on the knockouts: We were really surprised at our great results so far, and repeated the germination assay on our triple and quadruple mutants once again.
More work on the knockouts: We were really surprised at our great results so far, and repeated the germination assay on our triple and quadruple mutants once again.
3-7 September 2012
 All of our CotZ-GFP variants were examined with fluorescence microscopy! The brightest spores derived from our B 53 strain (containing Pcotyz-cotZrep-gfp-terminator).
All of our CotZ-GFP variants were examined with fluorescence microscopy! The brightest spores derived from our B 53 strain (containing Pcotyz-cotZrep-gfp-terminator).
 We worked more on the knockouts aspect of the GerminationSTOP module. We ran another germination assay for our triple and quadruple mutants. Plates of our spores diluted at 10-2, 10-4 and 10-6 show NO GERMINATION for our mutants, and plenty of germination for the WT168 positive control! We will try plating undiluted mutant spores to see if any germination occurs at higher concentrations.
We worked more on the knockouts aspect of the GerminationSTOP module. We ran another germination assay for our triple and quadruple mutants. Plates of our spores diluted at 10-2, 10-4 and 10-6 show NO GERMINATION for our mutants, and plenty of germination for the WT168 positive control! We will try plating undiluted mutant spores to see if any germination occurs at higher concentrations.
 More work on the Suicide switch! [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823042 PsspK], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823048 PspoIVB] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823041 PydfG] were cloned into pSB1C3 and verified by sequencing.
More work on the Suicide switch! [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823042 PsspK], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823048 PspoIVB] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823041 PydfG] were cloned into pSB1C3 and verified by sequencing.
PspoIVB-ecf41Bli aa1-204 were cloned into pSBBS4S and verified by sequencing.
PydfG/PsspK/PspoIVB were brought into pSBBS3C-luxABCDE.
 [http://partsregistry.org/Part:BBa_K823019 lacZ] was succesfully cloned into [http://partsregistry.org/Part:BBa_K823024 pSBBs4S-PXyl] and is functional (blue colonies with IPTG and X-Gal).
[http://partsregistry.org/Part:BBa_K823019 lacZ] was succesfully cloned into [http://partsregistry.org/Part:BBa_K823024 pSBBs4S-PXyl] and is functional (blue colonies with IPTG and X-Gal).
 A collection of useful tags in Freiburg standard and with RBS included was cloned into pSB1C3. The tags are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823034 3xFlag], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823035 HA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823036 cMyc], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823037 10xHis] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823038 Streptavidin].
A collection of useful tags in Freiburg standard and with RBS included was cloned into pSB1C3. The tags are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823034 3xFlag], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823035 HA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823036 cMyc], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823037 10xHis] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823038 Streptavidin].
August
26-31 August 2012
 Finally, we got our first glowing spores!! After 4 months of hard work, we have the first proof that this module works.
Finally, we got our first glowing spores!! After 4 months of hard work, we have the first proof that this module works.
 More on germination knockouts -- we created quadruple mutants using two variations on past mutants: cwlD::kan + cwlJ::spec + gerD::cm + sleB::mls and gerD::cm + sleB::mls + cwlJ::spec + cwlD::kan. See our Strains Collection.
Germination assay was performed on triple and quadruple mutants.
More on germination knockouts -- we created quadruple mutants using two variations on past mutants: cwlD::kan + cwlJ::spec + gerD::cm + sleB::mls and gerD::cm + sleB::mls + cwlJ::spec + cwlD::kan. See our Strains Collection.
Germination assay was performed on triple and quadruple mutants.
 For the Suicide switch, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823044 MazF] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823043 Ecf41Bli aa 1-204] were cloned into pSB1C3 and verified by sequencing.
For the Suicide switch, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823044 MazF] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823043 Ecf41Bli aa 1-204] were cloned into pSB1C3 and verified by sequencing.
 The BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 lacZ] for B. subtilis was shown to be functional in pSBBs0K-Pspac in E. coli and B. subtilis (blue color on plates with IPTG and X-Gal).
The BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 lacZ] for B. subtilis was shown to be functional in pSBBs0K-Pspac in E. coli and B. subtilis (blue color on plates with IPTG and X-Gal).
 The genes [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 luc+] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 mKate2], synthesized by GeneArt, were successfully cloned into pSB1C3 and sequenced.
The genes [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 luc+] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 mKate2], synthesized by GeneArt, were successfully cloned into pSB1C3 and sequenced.
20-24 August 2012
 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] with lacZα was finished and it works qualitatively.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] with lacZα was finished and it works qualitatively.
 Good and bad news this week. First the good news: Another big step towards the GFP-Sporobeads is done! We have the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823049 CotZ constructs] in pSBBS1C! Hopefully it will integrate easily! :D
Now the bad news: Finally we could start with the PcgeA evaluation! But contrary to our expectations, this promoter did not show any activity.
Good and bad news this week. First the good news: Another big step towards the GFP-Sporobeads is done! We have the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823049 CotZ constructs] in pSBBS1C! Hopefully it will integrate easily! :D
Now the bad news: Finally we could start with the PcgeA evaluation! But contrary to our expectations, this promoter did not show any activity.
13-17 August 2012
 For the knockouts, we used last week's double mutants to create triple mutants as follows: cwlD::kan + sleB::mls + cwlJ::spec ; cwlD::kan + sleB::mls + gerD::cm ; cwlD::kan + cwlJ::spec + gerD::cm ; and gerD::cm + sleB::mls + cwlJ::spec. See our Strains Collection.
For the knockouts, we used last week's double mutants to create triple mutants as follows: cwlD::kan + sleB::mls + cwlJ::spec ; cwlD::kan + sleB::mls + gerD::cm ; cwlD::kan + cwlJ::spec + gerD::cm ; and gerD::cm + sleB::mls + cwlJ::spec. See our Strains Collection.
 ß-glactosidase assay of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103] in pSBBs1C-lacZ in B. subtilis was performed.
The xylose-inducible promoter with the according repressor (which has a constitutive promoter, RBS and terminator) [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823015 xylR-PXyl] was cloned into pSB1C3 and sequenced.
ß-glactosidase assay of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103] in pSBBs1C-lacZ in B. subtilis was performed.
The xylose-inducible promoter with the according repressor (which has a constitutive promoter, RBS and terminator) [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823015 xylR-PXyl] was cloned into pSB1C3 and sequenced.
 At last Bacillus was keen on integrating the pSBBS3C-luxABCDE-PcgeA!! And the clean deletions of CotZ and CgeA worked out, too!
At last Bacillus was keen on integrating the pSBBS3C-luxABCDE-PcgeA!! And the clean deletions of CotZ and CgeA worked out, too!
6-10 August 2012
 Germination knockouts: We created four double-mutants using resistance-cassettes to knock out germination genes as follows: cwlD::kan + sleB::mls ; gerD::cm + sleB::mls ; gerD::cm + cwlD::kan ; cwlJ::spec + cwlD::kan. See our Strains Collection. Of these double mutants, we tested three strains in our germination assay. We also created the resistance cassette knockout cwlB::kan.
Germination knockouts: We created four double-mutants using resistance-cassettes to knock out germination genes as follows: cwlD::kan + sleB::mls ; gerD::cm + sleB::mls ; gerD::cm + cwlD::kan ; cwlJ::spec + cwlD::kan. See our Strains Collection. Of these double mutants, we tested three strains in our germination assay. We also created the resistance cassette knockout cwlB::kan.
 Finally, the last PstI site could be removed and [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE] was completed.
Also, the double terminator B0014 was cloned into pSB1C3.
Finally, the last PstI site could be removed and [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE] was completed.
Also, the double terminator B0014 was cloned into pSB1C3.
 The final Promoter-CotZ-GFP-Terminator constructs in pSB1C3 are ready!! Our Promoter-CgeAmut constructs are finally finished too.
The final Promoter-CotZ-GFP-Terminator constructs in pSB1C3 are ready!! Our Promoter-CgeAmut constructs are finally finished too.
July
23-27 July 2012
 This week is our Human Practice week!
From the 23rd to 25th of July we enjoyed the CAS SynBio conference and the visit of 10 iGEM teams!
This weekend, high school students participated in our Students Practical Course for three days and developed their ideas for useful Sporobeads!
It was a great week!
This week is our Human Practice week!
From the 23rd to 25th of July we enjoyed the CAS SynBio conference and the visit of 10 iGEM teams!
This weekend, high school students participated in our Students Practical Course for three days and developed their ideas for useful Sporobeads!
It was a great week!
 Plate reader measurements of the Bacillus promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI] and [http://partsregistry.org/Part:BBa_K823002 PlepA] are finished. Look at Data!
Plate reader measurements of the Bacillus promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI] and [http://partsregistry.org/Part:BBa_K823002 PlepA] are finished. Look at Data!
16-20 July 2012
 Not many of us are working on our modules this week, as we are helping out for the CAS SynBio conference and organising the Students Practical Course.
Not many of us are working on our modules this week, as we are helping out for the CAS SynBio conference and organising the Students Practical Course.
 Plate reader measurements of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823005 J23101], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106], [http://partsregistry.org/Part:BBa_K823009 J23107], [http://partsregistry.org/Part:BBa_K823010 J23113], [http://partsregistry.org/Part:BBa_K823011 J23114], [http://partsregistry.org/Part:BBa_K823012 J23115],
[http://partsregistry.org/Part:BBa_K823013 J23117], [http://partsregistry.org/Part:BBa_K823014 J23118] in the Bacillus reporter vector pSBBs3C-luxABCDE completed. Look at Data Anderson promoters!
Plate reader measurements of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823005 J23101], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106], [http://partsregistry.org/Part:BBa_K823009 J23107], [http://partsregistry.org/Part:BBa_K823010 J23113], [http://partsregistry.org/Part:BBa_K823011 J23114], [http://partsregistry.org/Part:BBa_K823012 J23115],
[http://partsregistry.org/Part:BBa_K823013 J23117], [http://partsregistry.org/Part:BBa_K823014 J23118] in the Bacillus reporter vector pSBBs3C-luxABCDE completed. Look at Data Anderson promoters!
 Cloning of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106] in the reporter vector pSBBs1C-lacZ for β-galactosidase assays finished.
Cloning of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106] in the reporter vector pSBBs1C-lacZ for β-galactosidase assays finished.
9-13 July 2012
 The vectors [http://partsregistry.org/Part:BBa_K823024 pSBBs4S-PXyl ], [http://partsregistry.org/Part:BBa_K823021 pSBBs1C-lacZ] and [http://partsregistry.org/Part:BBa_K823022 pSBBs4S ] were succesfully completed and tested by restriction digest as well as checked for red colony color.
The vectors [http://partsregistry.org/Part:BBa_K823024 pSBBs4S-PXyl ], [http://partsregistry.org/Part:BBa_K823021 pSBBs1C-lacZ] and [http://partsregistry.org/Part:BBa_K823022 pSBBs4S ] were succesfully completed and tested by restriction digest as well as checked for red colony color.
 We started with the clean deletions of CgeA and CotZ this week. It seems like it will take forever if you judge from the protocol length... What took even longer is the pSBBS3C-luxABCDE-PcgeA, but it is finished now!
We started with the clean deletions of CgeA and CotZ this week. It seems like it will take forever if you judge from the protocol length... What took even longer is the pSBBS3C-luxABCDE-PcgeA, but it is finished now!
2-6 July 2012
 In the knockouts part of the GerminationSTOP module, clean deletions of germination genes sleB and cwlB from PCR were accomplished. DNA was purified and frozen to be later transformed with Bacillus.
In the knockouts part of the GerminationSTOP module, clean deletions of germination genes sleB and cwlB from PCR were accomplished. DNA was purified and frozen to be later transformed with Bacillus.
 β-galactosidase assays of the promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823003 Pveg] are finished. Look at Data!
β-galactosidase assays of the promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823003 Pveg] are finished. Look at Data!
 Cloning of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823005 J23101], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106], [http://partsregistry.org/Part:BBa_K823009 J23107], [http://partsregistry.org/Part:BBa_K823010 J23113], [http://partsregistry.org/Part:BBa_K823011 J23114], [http://partsregistry.org/Part:BBa_K823012 J23115],
[http://partsregistry.org/Part:BBa_K823013 J23117], [http://partsregistry.org/Part:BBa_K823014 J23118] in pSB1C3 finished.
Cloning of the Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823005 J23101], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106], [http://partsregistry.org/Part:BBa_K823009 J23107], [http://partsregistry.org/Part:BBa_K823010 J23113], [http://partsregistry.org/Part:BBa_K823011 J23114], [http://partsregistry.org/Part:BBa_K823012 J23115],
[http://partsregistry.org/Part:BBa_K823013 J23117], [http://partsregistry.org/Part:BBa_K823014 J23118] in pSB1C3 finished.
 Our first plate reader experiments with Pcotyz and Pcotv are running! Like expected, they both show activity in the late stationary phase, thus during sporulation!
Our first plate reader experiments with Pcotyz and Pcotv are running! Like expected, they both show activity in the late stationary phase, thus during sporulation!
June
25-29 June 2012
 pSBBS3C-luxABCDE-Pcotyz and pSBBS3C-luxABCDE-Pcotv integrated into the B. subtilis genome!
Bad luck, we found an additional AgeI site in cgeA. It seems that this is the reason why we could not fuse gfp to it... Mutagenesis primers are designed and ordered!
pSBBS3C-luxABCDE-Pcotyz and pSBBS3C-luxABCDE-Pcotv integrated into the B. subtilis genome!
Bad luck, we found an additional AgeI site in cgeA. It seems that this is the reason why we could not fuse gfp to it... Mutagenesis primers are designed and ordered!
11-15 June 2012
 Knockouts: Clean deletions of germination genes cwlD, cwlJ, and gerD from PCR accomplished. DNA purified and frozen to be later transformed with Bacillus. Edit: the clean deletions were never used for transformation, but remain ready for future use.
Knockouts: Clean deletions of germination genes cwlD, cwlJ, and gerD from PCR accomplished. DNA purified and frozen to be later transformed with Bacillus. Edit: the clean deletions were never used for transformation, but remain ready for future use.
May
21-25 May 2012
 pSBBS3C-luxABCDE-Pcotyz and pSBBS3C-luxABCDE-Pcotv are ready for transformation into Bacillus!
pSBBS3C-luxABCDE-Pcotyz and pSBBS3C-luxABCDE-Pcotv are ready for transformation into Bacillus!
 Promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823003 Pveg] and [http://partsregistry.org/Part:BBa_K823002 PlepA] are now in the vector pSB1C3 as per the BioBrick standard for the registry.
Promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823003 Pveg] and [http://partsregistry.org/Part:BBa_K823002 PlepA] are now in the vector pSB1C3 as per the BioBrick standard for the registry.
 The Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823005 J23101], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106], [http://partsregistry.org/Part:BBa_K823009 J23107], [http://partsregistry.org/Part:BBa_K823010 J23113], [http://partsregistry.org/Part:BBa_K823011 J23114], [http://partsregistry.org/Part:BBa_K823012 J23115],
[http://partsregistry.org/Part:BBa_K823013 J23117], [http://partsregistry.org/Part:BBa_K823014 J23118] are now in the Bacillus reporter vector [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE] for measuring their activity as luminescence.
The Anderson promoters [http://partsregistry.org/Part:BBa_K823004 J23100], [http://partsregistry.org/Part:BBa_K823005 J23101], [http://partsregistry.org/Part:BBa_K823006 J23102], [http://partsregistry.org/Part:BBa_K823007 J23103], [http://partsregistry.org/Part:BBa_K823008 J23106], [http://partsregistry.org/Part:BBa_K823009 J23107], [http://partsregistry.org/Part:BBa_K823010 J23113], [http://partsregistry.org/Part:BBa_K823011 J23114], [http://partsregistry.org/Part:BBa_K823012 J23115],
[http://partsregistry.org/Part:BBa_K823013 J23117], [http://partsregistry.org/Part:BBa_K823014 J23118] are now in the Bacillus reporter vector [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE] for measuring their activity as luminescence.
 The Bacillus promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI] and [http://partsregistry.org/Part:BBa_K823002 PlepA] are in the Bacillus reporter vector [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE].
The Bacillus promoters [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI] and [http://partsregistry.org/Part:BBa_K823002 PlepA] are in the Bacillus reporter vector [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE].
14-18 May 2012
 Combining the BioBricks and integrating them into different vectors works well! But still there are constructs missing... We'll keep on working! The fusion of the up and down fragment of CotZ finally works!
Combining the BioBricks and integrating them into different vectors works well! But still there are constructs missing... We'll keep on working! The fusion of the up and down fragment of CotZ finally works!
7-11 May 2012
 The last weeks we tried to fuse the up and down fragment of our spore crust gene together for its clean deletion. For CgeA we have the first fused fragments! CotZ needs a little more attention. :)
The last weeks we tried to fuse the up and down fragment of our spore crust gene together for its clean deletion. For CgeA we have the first fused fragments! CotZ needs a little more attention. :)
 Cloning of [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823003 Pveg] and in vector [http://partsregistry.org/Part:BBa_K823021 pSBBs1C-lacZ] finished.
Cloning of [http://partsregistry.org/Part:BBa_K823000 PliaG], [http://partsregistry.org/Part:BBa_K823001 PliaI], [http://partsregistry.org/Part:BBa_K823003 Pveg] and in vector [http://partsregistry.org/Part:BBa_K823021 pSBBs1C-lacZ] finished.
April
30 April-4 May 2012
 We have the first essential BioBricks for this module: pSB1C3-CotZ, pSB1C3-PcotV, pSB1C3-PcotYZ, pSB1C3-PcgeA, pSB1C3-CgeA! Still working on the pSB1C3-GFP.
We have the first essential BioBricks for this module: pSB1C3-CotZ, pSB1C3-PcotV, pSB1C3-PcotYZ, pSB1C3-PcgeA, pSB1C3-CgeA! Still working on the pSB1C3-GFP.
23-27 April 2012
 pSBBs3C-luxABCDE with still one PstI site was created with RFP in the multile cloning site to have a vector for promoter measurments. edit: This PstI site was removed later and only that backbone is submitted to the registry.
pSBBs3C-luxABCDE with still one PstI site was created with RFP in the multile cloning site to have a vector for promoter measurments. edit: This PstI site was removed later and only that backbone is submitted to the registry.
16-20 April 2012
 The Sporobead team starts the work in the lab!
The Sporobead team starts the work in the lab!
 The cloning of the reporter vector [http://partsregistry.org/Part:BBa_K823021 pSBBs1C-lacZ] was finished.
The cloning of the reporter vector [http://partsregistry.org/Part:BBa_K823021 pSBBs1C-lacZ] was finished.
2-6 April 2012
 In our germination genes knockout work, we successfully created our first single knockouts of germination genes using a resistance cassettes: sleB::mls, gerD::cm and cwlJ::spec. See our Strains Collection.
We tried knocking out cwlB using the kan resistance cassette. Mutants of cwlB::kan grew very poorly.
In our germination genes knockout work, we successfully created our first single knockouts of germination genes using a resistance cassettes: sleB::mls, gerD::cm and cwlJ::spec. See our Strains Collection.
We tried knocking out cwlB using the kan resistance cassette. Mutants of cwlB::kan grew very poorly.
 With [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 pSBBs0K-Pspac ] our first vector for our Bacillus BioBrick Box was completed.
With [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 pSBBs0K-Pspac ] our first vector for our Bacillus BioBrick Box was completed.
March
26-30 March 2012
 Knockouts: We tried knocking out cwlB using the tet resistance cassette. Mutants of cwlB::tet grew very poorly.
Knockouts: We tried knocking out cwlB using the tet resistance cassette. Mutants of cwlB::tet grew very poorly.
19-23 March 2012
 For our germination gene knockouts, we decided which genes to knock out for the germination stop. Based on the work of [http://www.ncbi.nlm.nih.gov/pubmed/19554258 J. Kim and W. Schumann (2009)], we decided to knock out genes cwlB, gerD, cwlJ, and sleB. From the research of [http://www.ncbi.nlm.nih.gov/pubmed/11466293 B. Setlow et al (2001)], we also chose cwlD.
For our germination gene knockouts, we decided which genes to knock out for the germination stop. Based on the work of [http://www.ncbi.nlm.nih.gov/pubmed/19554258 J. Kim and W. Schumann (2009)], we decided to knock out genes cwlB, gerD, cwlJ, and sleB. From the research of [http://www.ncbi.nlm.nih.gov/pubmed/11466293 B. Setlow et al (2001)], we also chose cwlD.
5-9 March 2012
 We spent two full days (and nights) together planning our project in our "iGEM days." The idea of Beadzillus was born, and we got to know each other better as we divided the modules of the project, and learned each others' strengths and experience in different fields such as microbiology, ecology and biochemistry.
We spent two full days (and nights) together planning our project in our "iGEM days." The idea of Beadzillus was born, and we got to know each other better as we divided the modules of the project, and learned each others' strengths and experience in different fields such as microbiology, ecology and biochemistry.
February
Antibiotic abbreviation legend:
cm: chloramphenicol
kan: kanamycin
mls: Macrolide-Lincosamide-Streptogramin B
spec: spectinomycin
tet: tetracycline
 "
"