Team:Marburg SYNMIKRO/Project
From 2012.igem.org
(→Site-specific genetic recombination) |
(→Combinatorics) |
||
| (91 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
</html> | </html> | ||
| - | == | + | == General goal == |
| Line 26: | Line 26: | ||
</html> | </html> | ||
| - | == | + | == Playground == |
[[File:Syn_MR_Combinations.png|center|267px]] | [[File:Syn_MR_Combinations.png|center|267px]] | ||
| Line 38: | Line 38: | ||
</html> | </html> | ||
| - | == | + | == Combinatorics == |
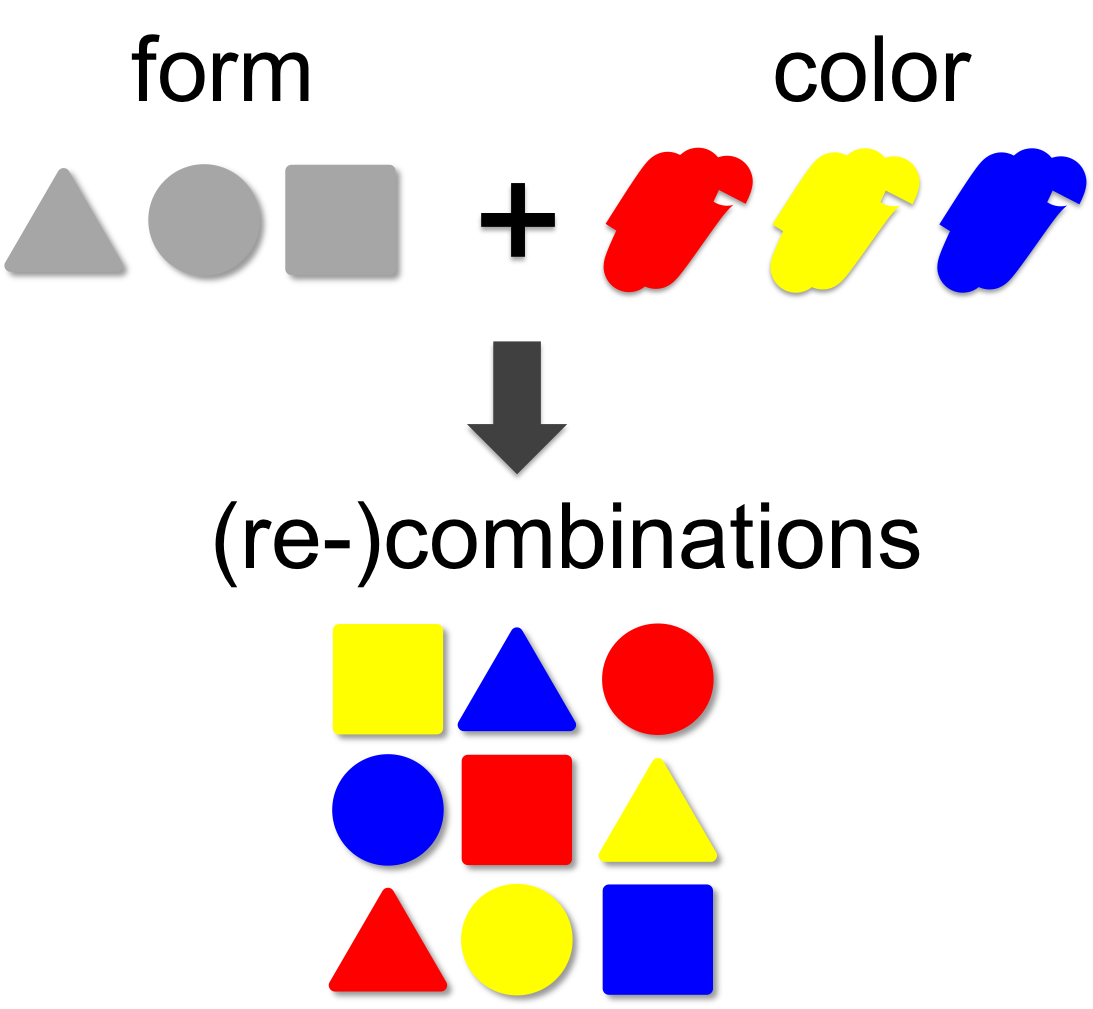
In mathematical theory the number of possible combinations results from the multiplication of elements. In our example (left) we combine three forms (triangle, square and circle) with three colors (red, blue and yellow). This results in 3 x 3 = 9 different combinations. | In mathematical theory the number of possible combinations results from the multiplication of elements. In our example (left) we combine three forms (triangle, square and circle) with three colors (red, blue and yellow). This results in 3 x 3 = 9 different combinations. | ||
| - | A practical example for the power of combinatorics is visualized on our [https://2012.igem.org/Team:Marburg_SYNMIKRO/ | + | A practical example for the power of combinatorics is visualized on our [https://2012.igem.org/Team:Marburg_SYNMIKRO/TeamProfile team page], where you can enlarge the size of the iGEM team by generating additional team members through recombination of the existing ones. By this method a total of 14 x 14 x 14 = '''2,744''' novel mixed-up team members can be designed. |
<html> | <html> | ||
</div> | </div> | ||
| Line 49: | Line 49: | ||
</html> | </html> | ||
| - | == | + | == VDJ-Recombination == |
| - | Our "Recombinator" system for the generation of novel fusion proteins in vivo was inspired by the generation of antibody diversity | + | Our "Recombinator" system for the generation of novel fusion proteins in vivo was inspired by the generation of antibody diversity in the adaptive immune system. The enormous number of different antibodies is generated by recombination of a limited number of segments that are recombined to form functional antibody chains. This occurs in the B-cells of the immune system and is essential for the recognition of specific antigens. |
| - | + | <br/> | |
| + | <br/> | ||
[[File:Syn_MR_VDJ-recombination.png|center|850px]] | [[File:Syn_MR_VDJ-recombination.png|center|850px]] | ||
| - | + | <br/> | |
Antibodies consist of two heavy and two light chains which are covalently connected by disulfide bonds. Both, the heavy and the light chain can be subdivided into constant and variable regions. The variable parts confer the ability to recognize specific antigens. During our life, the immune system is confronted with thousands of different antigens from bacteria, viruses and other microorganisms. This raises the question how antibody diversity is generated during development. | Antibodies consist of two heavy and two light chains which are covalently connected by disulfide bonds. Both, the heavy and the light chain can be subdivided into constant and variable regions. The variable parts confer the ability to recognize specific antigens. During our life, the immune system is confronted with thousands of different antigens from bacteria, viruses and other microorganisms. This raises the question how antibody diversity is generated during development. | ||
| Line 70: | Line 71: | ||
</html> | </html> | ||
| - | == | + | == Site-specific recombination == |
| - | Genetic recombination is essential to maintain the integrity of large genomes. Double strand breaks in the DNA are repaired by the process of homologous recombination which is conserved from bacteria to man. During sexual propagation homologous recombination creates genetic diversity by | + | Genetic recombination is essential to maintain the integrity of large genomes. Double strand breaks in the DNA are repaired by the process of homologous recombination which is conserved from bacteria to man. During sexual propagation homologous recombination occurs during meiosis and creates genetic diversity by exchanging DNA segments between chromosomes. |
| + | <br/> | ||
<br/> | <br/> | ||
[[File:Syn_MR_Inversion_Deletion.png|center|400px]] | [[File:Syn_MR_Inversion_Deletion.png|center|400px]] | ||
| - | + | <br/> | |
| - | In addition to homologus recombination several site-specific DNA recombination systems have | + | In addition to homologus recombination, several site-specific DNA recombination systems have evolved. Site-specific recombination is used for integration of phage lambda, phase variation of pathogenic ''Salmonella'' strains, for VDJ-recombination during antibody development and for several other biological processes, which involve specific DNA rearrangements. In all these cases, specialized enzymes recognize short DNA sequences flanking a segment of DNA. Recombination occurs by cutting and religating these short elements. Depending on the orientation of recombination sites, this process either results in INVERSION or DELETION of the intervening DNA segment. If the recombination sites are arranged as inverted repeats, the segment is inverted; if they form direct repeats, the intervening segment will be removed by recombination. |
<html> | <html> | ||
</div> | </div> | ||
| Line 84: | Line 86: | ||
</html> | </html> | ||
| - | == | + | == Phage Mu == |
| - | The bacteriophage Mu is a temperate virus which has an unusual lifestyle. | + | The bacteriophage Mu is a temperate bacterial virus which has an unusual lifestyle. During lytic phase it propagates in the bacterial host cell by replicative transposition. During lysogenic phase it also behaves like a transposon and integrates into the host genome at a random position. This results in the occurrence of genetic mutations and hence the phage got its name ("'''mu'''tator phage"). This has been used to construct small derivatives (mini Mu) which can be used for tagging mutagenesis. |
| - | + | <br/> | |
| + | <br/> | ||
| + | [[File:Syn_MR_PhageMu_small.png|center|402px]] | ||
| + | <br/> | ||
| + | Interestingly, phage Mu has a broad host range and can infect very different types of bacteria. Recognition of different hosts by the phage depends on the expression of specific tail proteins (S and U) which are located in the phage genome on the invertible G-segment. Dependent on the orientation of the G-segment, G(+) or G(-), different versions of the tail proteins S and U are expressed and recognize different lipopolysaccharides in the cell wall of bacteria. | ||
<html> | <html> | ||
| Line 95: | Line 101: | ||
<div class="conboxs"> | <div class="conboxs"> | ||
</html> | </html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | == Gin recombination == | ||
| + | The invertible G-segment of bacteriophage Mu contains two genes, S and U, which are transcribed from a common promoter located outside the G-segment. Depending on its orientation either S and U or S' and U' are expressed. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | [[File:Syn_MR_G-segment.png|center|440px]] | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Inversion of the G-segment is catalyzed by the site-specific DNA recombinase Gin which is encoded on the phage genome. The ''gin'' gene is located adjacent to the G-segment. Gin invertase binds to the short gixL and gixR recombination-sites flanking the G-segment. Recombination between these inverted sites results in inversion of the G-segment. | ||
<html> | <html> | ||
| Line 109: | Line 118: | ||
<div class="conboxs"> | <div class="conboxs"> | ||
</html> | </html> | ||
| - | == | + | |
| - | + | == Enhancer == | |
| - | Site-specific recombination catalyzed by Gin recombinase requires the formation of a synaptonemal complex with aligned recombination sites (below). | + | Site-specific recombination catalyzed by Gin recombinase requires the formation of a synaptonemal DNA-protein complex with aligned recombination sites (below). |
| + | [[File:Syn_MR_Enhancer.png|center|215px]] | ||
| + | Efficient recombination occurs only in the presence of a recombinational enhancer (open rectangle) which is located within the inverted DNA segment. The enhancer is bound by the bacterial protein FIS, which stimulates formation of the synaptonemal complex. | ||
<html> | <html> | ||
</div> | </div> | ||
| Line 120: | Line 131: | ||
</html> | </html> | ||
| - | == | + | == Construct == |
| - | + | ˈThe Recombinatorˈ is able to recombine diverse protein domains resulting in expression of chimeric proteins. The system is based on the site-specific Gin recombination system of bacteriophage Mu. Our aim was to generate random fusions of two different modules. To prevent "over"-recombination which would result in deletion of all modules but one, we inserted a recombinational enhancer between the different modules. Since efficient recombination between two '''gixR''' sites (yellow triangles) depends on the presence of the '''enhancer''' (Klippel et al., 1993) recombination stops immediately after removal of the central segment. This guarantees that our system generates a random fusion of one A-module with one B module. | |
| - | ˈThe Recombinatorˈ is able to recombine diverse protein | + | |
| - | |||
| - | + | [[File:Syn_MR_construct_alt.png|center|875px]] | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | To demonstrate the proof-of-principle of the recombinator, we used bacterial intracellular localization domains as A-modules and fluorescent proteins as B-modules. The A-modules were taken from the bacterial HU, GroES and Amidase C proteins. These proteins are targeted to the cell pole, the periplasm and the nucleoid, respectively (Bernhardt et al., 2003; Wery et al., 2001; Li et al., 2012). Each A-module is preceded by a constant promoter ([http://partsregistry.org/Part:BBa_J23108 BBa_J23108]) and a ribosomal binding site ([http://partsregistry.org/Part:BBa_J61100 BBa_J61100]) and is followed by a Gin recombination site (gixR). To prevent accumulation of the protein domains used for localization, we have placed a transcription terminator downstream of the A-modules such that the resulting transcripts contain no stop codon. Due to missing stop codons in the A-modules the ribosomes are stalled at the mRNA. Such stalled ribosomes are recognized in bacteria by tmRNA (transfer-messenger-RNA). This results in the recycling of the ribosomes and in the degradation of the tagged protein by proteases (see review by [http://www.ncbi.nlm.nih.gov/pubmed/18557701 Keiler (2008)] for further details). | |
| - | + | <br/> | |
| - | + | <br/> | |
| - | + | [[File:Syn_MR_Fusionproduct.png|center|600px]] | |
| - | + | <br/> | |
| - | + | <br/> | |
| - | + | After recombination, one of the possible fusion products will be stably expressed from the promoter since now the transcript is terminated after the B-module and thus contains a stop codon. Gin recombinase is provided by an extra plasmid which expresses Gin under control of the IPTG-inducible Plac promoter (BBa_). Induction of Gin recombinase will trigger recombination between ''gixR'' sites. For instance: A2 will be fused to B2 by recombination, then A3, the enhancer, the sacB gene and B1 are deleted. The resulting fusion protein A2-B2 is supposed to be found at the nucleoid in the middle of the cells and to fluoresce red. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | To select for plasmids in which productive recombination events have occurred, we used the toxic ''sacB'' gene for counter-selection. The ''Bacillus subtilis'' ''sacB'' gene codes for the secreted enzyme levansucrase which belongs to the glycosyltransferase family. The enzyme uses sucrose as substrate to produce the <html>polymeric levan (2,6-β-D-fructosyl)<sub>n</sub></html>. Levan has an lethal effect on cells of gram negative bacteria like ''Escherichia coli'' ([http://www.ncbi.nlm.nih.gov/pubmed/2997137 Gay et al., 1985]). For selection of recombined modules cells will be grown in the presence of sucrose. | ||
| + | The usage of three A- and B-modules results in only nine possible combinations. But the number of possible fusion proteins can be scaled up by utilization of further domains. | ||
<html> | <html> | ||
| Line 164: | Line 157: | ||
</html> | </html> | ||
| - | == | + | == Localization domains == |
| - | + | We have screened the literature and identified several bacterial proteins with particular localization within the cell. We decided to use GroES for localization at the cell poles, the small DNA binding protein HU to mark the nucleoid and the amidase AmiC, which is targeted to the periplasm. Depicted are micrographs from the articles by Li ''et al.'', 2012, Wery ''et al.'', 2001 and Bernhardt ''et al.'', 2004. | |
| - | We have screened the literature and identified several bacterial proteins with particular localization within the cell. We decided to use GroES for localization at the cell poles, the small DNA binding protein HU to mark the nucleoid and the amidase AmiC, which is targeted to the periplasm. Depicted are micrographs from the articles by Li ''et al.'', 2012, Wery ''et al.'', 2001 and | + | <br/> |
| - | + | <br/> | |
| - | + | [[File:Syn_MR_Localization_Domains.png|center|435px]] | |
| - | + | <br/> | |
| - | + | <br/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | HU belongs to the group of small ˈhistone-likeˈ DNA-binding proteins and has its function in the spatial organization of genomic DNA ([http://www.ncbi.nlm.nih.gov/pubmed/11278069 Wery et al., 2001]). B-modules fused with this domain are expected to accumulate in the nucleoid. Furthermore we used the chaperonin GroES which localizes at the cell pole. GroES interacts with GroEL and assists protein folding in the presence of ATP ([http://www.ncbi.nlm.nih.gov/pubmed/22380631 Li et al., 2012]). The third domain was taken from the cell wall degrading enzyme Amidase C. Amidase C operates during cell division and is localized in the periplasm ([http://www.ncbi.nlm.nih.gov/pubmed/12787347 Bernhardt et al., 2004]). | ||
<html> | <html> | ||
</div> | </div> | ||
| Line 187: | Line 174: | ||
</html> | </html> | ||
| - | == | + | == Fluorescent proteins == |
| - | + | Fluorescent proteins have been established as an invaluable tool to study the intracellular localization of proteins. They were first discovered in the jellyfish ''Aequorea victoria''. Later, fluorescent proteins were also found in other systems and in many other colors. The discovery of GFP was honored in 2008 by the [http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2008/ Nobel prize] to Martin Chalfie, Osamu Shimomura, and Roger Y. Tsien. | |
| - | + | <br/> | |
| - | [http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2008/ Nobel prize] | + | <br/> |
| - | + | [[File:Syn_MR_iGESM.jpg|center|347px|mRFP on plate]] | |
| + | <br/> | ||
| + | <br/> | ||
| + | We have used green (GFP), red (mRFP) and blue (CFP) variants of fluorescent proteins. In the registry, only one version of GFP was present as biobrick ([http://partsregistry.org/Part:BBa_K125500 BBa_K125500]), which is designed for fusion proteins. All other biobricks contain a stop codon in the prefix which is maintained in the scar. Therefore, we improved the existing mRFP and CFP biobricks by converting them into fusion-ready constructs. | ||
<html> | <html> | ||
</div> | </div> | ||
</html> | </html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
| Line 219: | Line 192: | ||
</html> | </html> | ||
| - | == | + | == Outlook == |
| - | + | Our Recombinator imitates the process of random shuffling of protein domains, which has been proposed to be a major force during evolution ([http://www.ncbi.nlm.nih.gov/pubmed/7574483 Doolittle, 1995]). Modified enzymes created by combinatorial fusion in vivo can reveal a higher specificity to their substrates or increase their activities to an extended range of substrates. Moreover, proteins can acquire improved functions like alternative partners for interaction or altered stability. Furthermore our system does not depend on rational design that requires knowledge of the three-dimensional structure of the proteins. | |
| - | + | Besides industrial applications such as the optimization of cellulases, lipases, amylases or the production of synthetic chemicals in novel metabolic pathways the development and improvement of pharmaceuticals and antibiotics attracts attention. The discovery of potential useful pharmaceuticals may be facilitated by the generation of combinatorial protein domain libraries provided by our approach. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html> | <html> | ||
</div> | </div> | ||
</html> | </html> | ||
| + | |||
<html> | <html> | ||
| Line 238: | Line 204: | ||
</html> | </html> | ||
| - | == | + | == References == |
| - | + | Bernhardt TG and de Boer PA (2003) The ''Escherichia coli'' amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. ''Mol Microbiol'' '''48''':1171-82. ([http://www.ncbi.nlm.nih.gov/pubmed/12787347 pubmed]) | |
| - | + | Doolittle RF (1995) The multiplicity of domains in proteins. ''Annu Rev Biochem'' '''64''':287-314. ([http://www.ncbi.nlm.nih.gov/pubmed/7574483 pubmed]) | |
| - | + | Faelen M, Resibois A and Toussaint A (1979) Mini-mu: an insertion element derived from temperate phage mu-1. ''Cold Spring Harb Symp Quant Biol'' '''43''':1169-77. ([http://www.ncbi.nlm.nih.gov/pubmed/158471 pubmed]) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Gay P, Le Coq D, Steinmetz M, Berkelman T and Kado CI (1985) Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. ''J Bacteriol'' '''164''':918–921. ([http://www.ncbi.nlm.nih.gov/pubmed/2997137 pubmed]) | |
| - | + | Keiler KC (2008) Biology of trans-translation. ''Annu Rev Microbiol'' '''62''':133-51. ([http://www.ncbi.nlm.nih.gov/pubmed/18557701 pubmed]) | |
| - | + | Klippel A, Kanaar R, Kahmann R and Cozzarelli NR (1993) Analysis of strand exchange and DNA binding of enhancer-independent Gin recombinase mutants. ''EMBO J'' '''12''':1047-1057. ([http://www.ncbi.nlm.nih.gov/pubmed/8384550 pubmed]) | |
| - | + | Li G and Young KD (2012) Isolation and identification of new inner membrane-associated proteins that localize to cell poles in ''Escherichia coli''. ''Mol Microbiol'' '''84''':276–295. ([http://www.ncbi.nlm.nih.gov/pubmed/22380631 pubmed]) | |
| + | Wery M, Woldringh CL and Rouviere-Yaniv J (2001) HU-GFP and DAPI co-localize on the ''Escherichia coli'' nucleoid. ''Biochimie'' '''83''':193-200. ([http://www.ncbi.nlm.nih.gov/pubmed/11278069 pubmed]) | ||
{{:Team:Marburg_SYNMIKRO:Template:Footer}} | {{:Team:Marburg_SYNMIKRO:Template:Footer}} | ||
Latest revision as of 09:59, 19 October 2012

Contents |
Summary
We aimed to establish a system in Escherichia coli cells that generates a large number of novel proteins by combinatorial fusion of subfragments. Our project was inspired by the VDJ-recombination of the vertebrate immune system. There, a limited number of protein coding sequences is used to generate the enormous diversity of antibodies. We designed A and B modules containing different protein coding subfragments to be recombined. Fusion of one A fragment with one B fragment is achieved by the site-specific DNA recombinase Gin of bacteriophage Mu. Recombination sites were arranged as direct repeats thus causing deletions upon recombination. Gin catalyzed recombination depends on the presence of an enhancer element.This enhancer was placed together with the suicide gene sacB between the A and B modules. This design guarantees that recombination automatically stops when the central fragment is deleted. To test our system and to visualize the combinatorial activity we fused fluorescent proteins of different color to different intracellular localization domains. We expect that after successful recombination E. coli cells will glow in different colors located at different positions within the cell.
General goal
The development of novel proteins and the Improvement of existing ones by genetic engineering has a tremendous impact on our daily lives. We all benefit from advances in this field be it for medical, industrial or environmental applications. The central goal of our project is the random generation of novel protein variants. The Marburg_SYNMIKRO team 2012 has created ˈThe Recombinatorˈ: an intelligent Genetically Engineered Slot Machine (iGESM). "The Recombinator" can be used to produce large numbers of novel proteins by combinatorial fusion of functional domains.
Combinatorics
In mathematical theory the number of possible combinations results from the multiplication of elements. In our example (left) we combine three forms (triangle, square and circle) with three colors (red, blue and yellow). This results in 3 x 3 = 9 different combinations. A practical example for the power of combinatorics is visualized on our team page, where you can enlarge the size of the iGEM team by generating additional team members through recombination of the existing ones. By this method a total of 14 x 14 x 14 = 2,744 novel mixed-up team members can be designed.
VDJ-Recombination
Our "Recombinator" system for the generation of novel fusion proteins in vivo was inspired by the generation of antibody diversity in the adaptive immune system. The enormous number of different antibodies is generated by recombination of a limited number of segments that are recombined to form functional antibody chains. This occurs in the B-cells of the immune system and is essential for the recognition of specific antigens.
Antibodies consist of two heavy and two light chains which are covalently connected by disulfide bonds. Both, the heavy and the light chain can be subdivided into constant and variable regions. The variable parts confer the ability to recognize specific antigens. During our life, the immune system is confronted with thousands of different antigens from bacteria, viruses and other microorganisms. This raises the question how antibody diversity is generated during development.
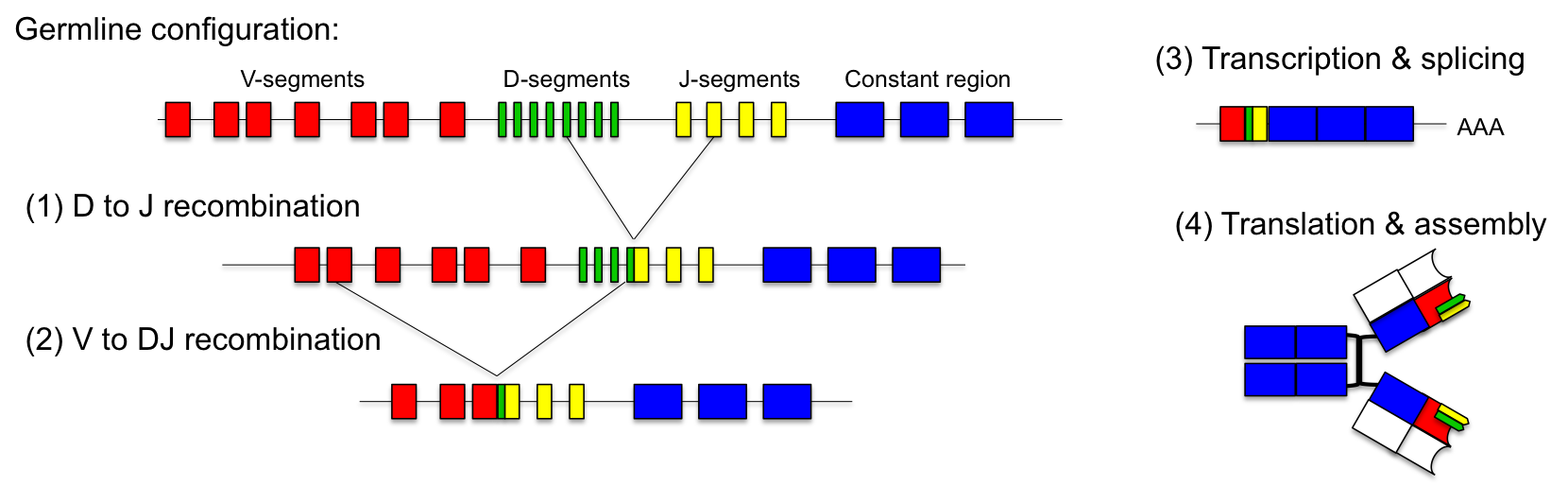
Nature has invented an ingenious solution to this problem: the combinatorial design of the variable regions of the antibodies. In the germline, the genetic locus of the light chain consists of up to 40 V-segments and 5 J-segments. The heavy chain contains 50 V-, 27 D- and 6 J-segments. During maturation of the immune system antibodies are generated by random fusion of these segments.This process is called VDJ-recombination and is catalyzed by the DNA recombinases (RAG-1 and RAG-2).
For the light chain, one V-segment gets fused to one of the J-segments by recombination. The production of the variable heavy chain occurs in two steps. Initially, one of the J-segments is fused to the D-segment (1). Then, the combined DJ-sequence is added to one of the V-segments (2). The recombined antibody gene is transcribed together with the constant part of the antibody, which is added as exon by splicing (3). Heavy and light chains are assembled into functional antibodies (4). Stability of the secreted antibodies is reached by covalent disulfide bonds between heavy and light chains. Since each antibody is generated by combinatorial fusion of one of 40 V-segments, 5 J-segments for the light chain and one of 50 V-, 27 D- and 6 J-segments for the heavy chain, the number of possible combinations amounts to 1.6 x 106. Furthermore, during the recombination process additional mutations are introduced. This increases the number of possible rearrangements to more than 1010.
Site-specific recombination
Genetic recombination is essential to maintain the integrity of large genomes. Double strand breaks in the DNA are repaired by the process of homologous recombination which is conserved from bacteria to man. During sexual propagation homologous recombination occurs during meiosis and creates genetic diversity by exchanging DNA segments between chromosomes.
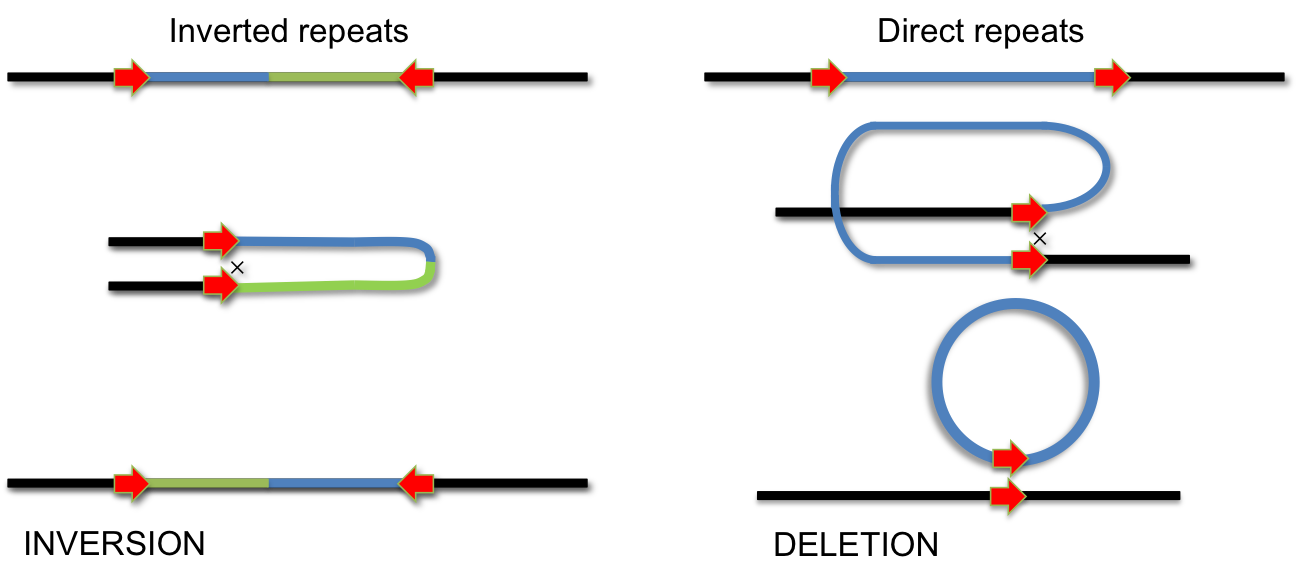
In addition to homologus recombination, several site-specific DNA recombination systems have evolved. Site-specific recombination is used for integration of phage lambda, phase variation of pathogenic Salmonella strains, for VDJ-recombination during antibody development and for several other biological processes, which involve specific DNA rearrangements. In all these cases, specialized enzymes recognize short DNA sequences flanking a segment of DNA. Recombination occurs by cutting and religating these short elements. Depending on the orientation of recombination sites, this process either results in INVERSION or DELETION of the intervening DNA segment. If the recombination sites are arranged as inverted repeats, the segment is inverted; if they form direct repeats, the intervening segment will be removed by recombination.
Phage Mu
The bacteriophage Mu is a temperate bacterial virus which has an unusual lifestyle. During lytic phase it propagates in the bacterial host cell by replicative transposition. During lysogenic phase it also behaves like a transposon and integrates into the host genome at a random position. This results in the occurrence of genetic mutations and hence the phage got its name ("mutator phage"). This has been used to construct small derivatives (mini Mu) which can be used for tagging mutagenesis.
Interestingly, phage Mu has a broad host range and can infect very different types of bacteria. Recognition of different hosts by the phage depends on the expression of specific tail proteins (S and U) which are located in the phage genome on the invertible G-segment. Dependent on the orientation of the G-segment, G(+) or G(-), different versions of the tail proteins S and U are expressed and recognize different lipopolysaccharides in the cell wall of bacteria.
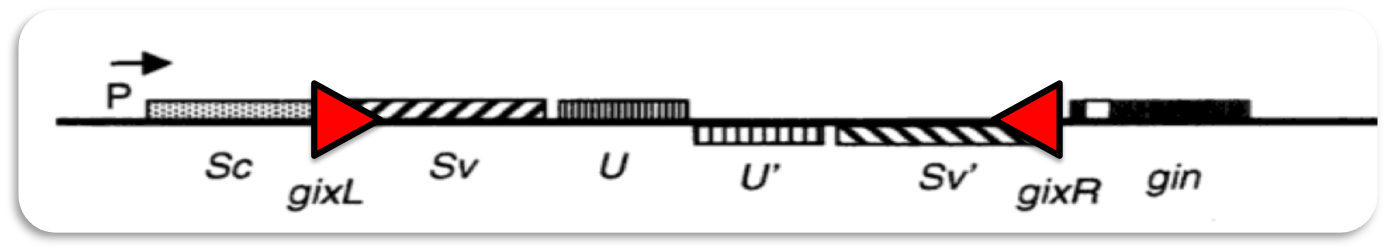
Gin recombination
The invertible G-segment of bacteriophage Mu contains two genes, S and U, which are transcribed from a common promoter located outside the G-segment. Depending on its orientation either S and U or S' and U' are expressed.
Inversion of the G-segment is catalyzed by the site-specific DNA recombinase Gin which is encoded on the phage genome. The gin gene is located adjacent to the G-segment. Gin invertase binds to the short gixL and gixR recombination-sites flanking the G-segment. Recombination between these inverted sites results in inversion of the G-segment.
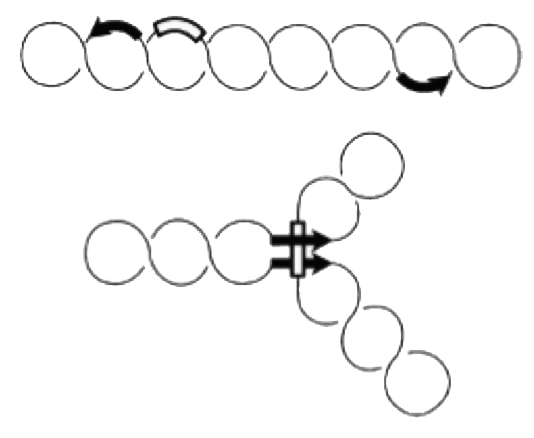
Enhancer
Site-specific recombination catalyzed by Gin recombinase requires the formation of a synaptonemal DNA-protein complex with aligned recombination sites (below).
Efficient recombination occurs only in the presence of a recombinational enhancer (open rectangle) which is located within the inverted DNA segment. The enhancer is bound by the bacterial protein FIS, which stimulates formation of the synaptonemal complex.
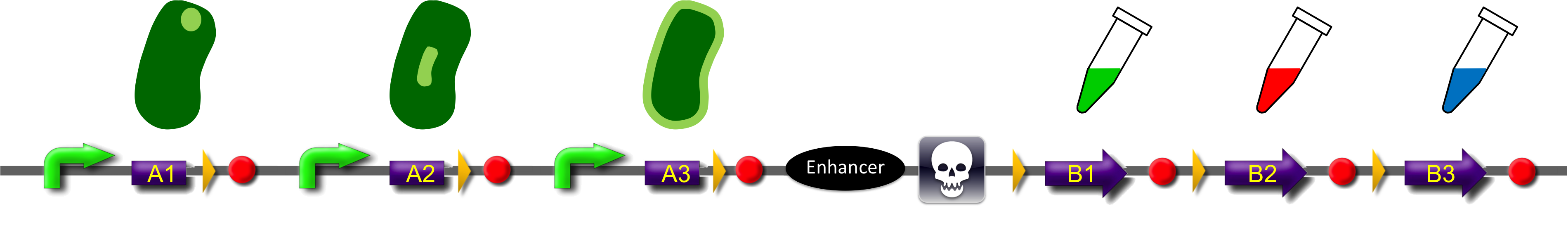
Construct
ˈThe Recombinatorˈ is able to recombine diverse protein domains resulting in expression of chimeric proteins. The system is based on the site-specific Gin recombination system of bacteriophage Mu. Our aim was to generate random fusions of two different modules. To prevent "over"-recombination which would result in deletion of all modules but one, we inserted a recombinational enhancer between the different modules. Since efficient recombination between two gixR sites (yellow triangles) depends on the presence of the enhancer (Klippel et al., 1993) recombination stops immediately after removal of the central segment. This guarantees that our system generates a random fusion of one A-module with one B module.
To demonstrate the proof-of-principle of the recombinator, we used bacterial intracellular localization domains as A-modules and fluorescent proteins as B-modules. The A-modules were taken from the bacterial HU, GroES and Amidase C proteins. These proteins are targeted to the cell pole, the periplasm and the nucleoid, respectively (Bernhardt et al., 2003; Wery et al., 2001; Li et al., 2012). Each A-module is preceded by a constant promoter ([http://partsregistry.org/Part:BBa_J23108 BBa_J23108]) and a ribosomal binding site ([http://partsregistry.org/Part:BBa_J61100 BBa_J61100]) and is followed by a Gin recombination site (gixR). To prevent accumulation of the protein domains used for localization, we have placed a transcription terminator downstream of the A-modules such that the resulting transcripts contain no stop codon. Due to missing stop codons in the A-modules the ribosomes are stalled at the mRNA. Such stalled ribosomes are recognized in bacteria by tmRNA (transfer-messenger-RNA). This results in the recycling of the ribosomes and in the degradation of the tagged protein by proteases (see review by [http://www.ncbi.nlm.nih.gov/pubmed/18557701 Keiler (2008)] for further details).
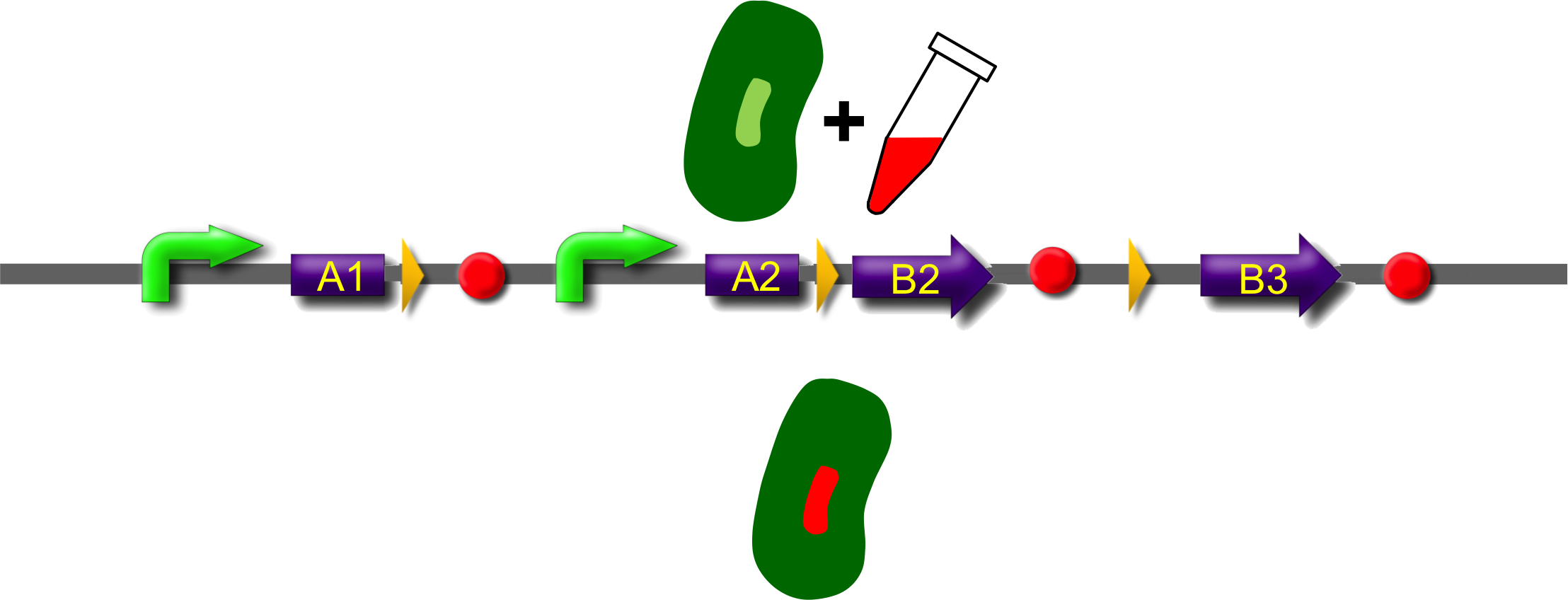
After recombination, one of the possible fusion products will be stably expressed from the promoter since now the transcript is terminated after the B-module and thus contains a stop codon. Gin recombinase is provided by an extra plasmid which expresses Gin under control of the IPTG-inducible Plac promoter (BBa_). Induction of Gin recombinase will trigger recombination between gixR sites. For instance: A2 will be fused to B2 by recombination, then A3, the enhancer, the sacB gene and B1 are deleted. The resulting fusion protein A2-B2 is supposed to be found at the nucleoid in the middle of the cells and to fluoresce red.
To select for plasmids in which productive recombination events have occurred, we used the toxic sacB gene for counter-selection. The Bacillus subtilis sacB gene codes for the secreted enzyme levansucrase which belongs to the glycosyltransferase family. The enzyme uses sucrose as substrate to produce the polymeric levan (2,6-β-D-fructosyl)n. Levan has an lethal effect on cells of gram negative bacteria like Escherichia coli ([http://www.ncbi.nlm.nih.gov/pubmed/2997137 Gay et al., 1985]). For selection of recombined modules cells will be grown in the presence of sucrose. The usage of three A- and B-modules results in only nine possible combinations. But the number of possible fusion proteins can be scaled up by utilization of further domains.
Localization domains
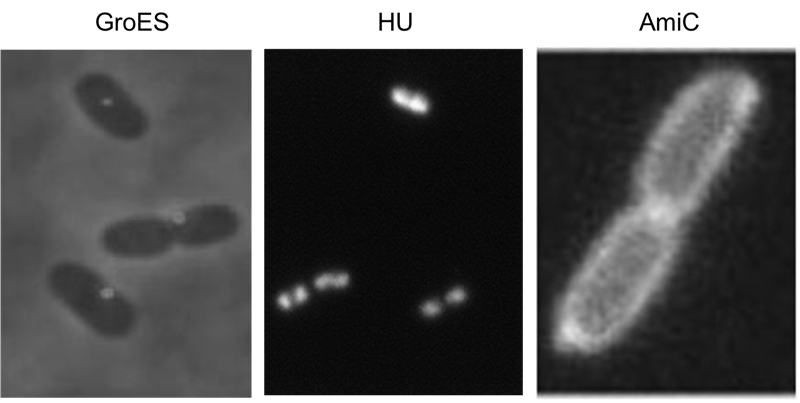
We have screened the literature and identified several bacterial proteins with particular localization within the cell. We decided to use GroES for localization at the cell poles, the small DNA binding protein HU to mark the nucleoid and the amidase AmiC, which is targeted to the periplasm. Depicted are micrographs from the articles by Li et al., 2012, Wery et al., 2001 and Bernhardt et al., 2004.
HU belongs to the group of small ˈhistone-likeˈ DNA-binding proteins and has its function in the spatial organization of genomic DNA ([http://www.ncbi.nlm.nih.gov/pubmed/11278069 Wery et al., 2001]). B-modules fused with this domain are expected to accumulate in the nucleoid. Furthermore we used the chaperonin GroES which localizes at the cell pole. GroES interacts with GroEL and assists protein folding in the presence of ATP ([http://www.ncbi.nlm.nih.gov/pubmed/22380631 Li et al., 2012]). The third domain was taken from the cell wall degrading enzyme Amidase C. Amidase C operates during cell division and is localized in the periplasm ([http://www.ncbi.nlm.nih.gov/pubmed/12787347 Bernhardt et al., 2004]).
Fluorescent proteins
Fluorescent proteins have been established as an invaluable tool to study the intracellular localization of proteins. They were first discovered in the jellyfish Aequorea victoria. Later, fluorescent proteins were also found in other systems and in many other colors. The discovery of GFP was honored in 2008 by the [http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2008/ Nobel prize] to Martin Chalfie, Osamu Shimomura, and Roger Y. Tsien.
We have used green (GFP), red (mRFP) and blue (CFP) variants of fluorescent proteins. In the registry, only one version of GFP was present as biobrick ([http://partsregistry.org/Part:BBa_K125500 BBa_K125500]), which is designed for fusion proteins. All other biobricks contain a stop codon in the prefix which is maintained in the scar. Therefore, we improved the existing mRFP and CFP biobricks by converting them into fusion-ready constructs.
Outlook
Our Recombinator imitates the process of random shuffling of protein domains, which has been proposed to be a major force during evolution ([http://www.ncbi.nlm.nih.gov/pubmed/7574483 Doolittle, 1995]). Modified enzymes created by combinatorial fusion in vivo can reveal a higher specificity to their substrates or increase their activities to an extended range of substrates. Moreover, proteins can acquire improved functions like alternative partners for interaction or altered stability. Furthermore our system does not depend on rational design that requires knowledge of the three-dimensional structure of the proteins. Besides industrial applications such as the optimization of cellulases, lipases, amylases or the production of synthetic chemicals in novel metabolic pathways the development and improvement of pharmaceuticals and antibiotics attracts attention. The discovery of potential useful pharmaceuticals may be facilitated by the generation of combinatorial protein domain libraries provided by our approach.
References
Bernhardt TG and de Boer PA (2003) The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48:1171-82. ([http://www.ncbi.nlm.nih.gov/pubmed/12787347 pubmed])
Doolittle RF (1995) The multiplicity of domains in proteins. Annu Rev Biochem 64:287-314. ([http://www.ncbi.nlm.nih.gov/pubmed/7574483 pubmed])
Faelen M, Resibois A and Toussaint A (1979) Mini-mu: an insertion element derived from temperate phage mu-1. Cold Spring Harb Symp Quant Biol 43:1169-77. ([http://www.ncbi.nlm.nih.gov/pubmed/158471 pubmed])
Gay P, Le Coq D, Steinmetz M, Berkelman T and Kado CI (1985) Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol 164:918–921. ([http://www.ncbi.nlm.nih.gov/pubmed/2997137 pubmed])
Keiler KC (2008) Biology of trans-translation. Annu Rev Microbiol 62:133-51. ([http://www.ncbi.nlm.nih.gov/pubmed/18557701 pubmed])
Klippel A, Kanaar R, Kahmann R and Cozzarelli NR (1993) Analysis of strand exchange and DNA binding of enhancer-independent Gin recombinase mutants. EMBO J 12:1047-1057. ([http://www.ncbi.nlm.nih.gov/pubmed/8384550 pubmed])
Li G and Young KD (2012) Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol Microbiol 84:276–295. ([http://www.ncbi.nlm.nih.gov/pubmed/22380631 pubmed])
Wery M, Woldringh CL and Rouviere-Yaniv J (2001) HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie 83:193-200. ([http://www.ncbi.nlm.nih.gov/pubmed/11278069 pubmed])
 "
"