Team:Potsdam Bioware/Project/Part Virus
From 2012.igem.org
K.tomschek (Talk | contribs) (→Discussion) |
(→Targeting the virus against the advanced antibody construct) |
||
| (166 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class="box_round gradient_grey"> | <div class="box_round gradient_grey"> | ||
| - | ''' | + | ==Selection Module== |
| - | + | ||
| + | ===Abstract Selection Module=== | ||
| + | |||
| + | The "selection module" comprises a set of methods to select the desired cell clones from the collection of cells each expressing one variant of the antibody construct. Depending on the needs, different selection methods might have specific advantages. We set up three methods, whereby two are well established but discontinuous: i) fluorescent activated cell sorting (FACS), which yields many data but requires expensive instrumentation and ii) magnetic beads, which are less expensive and easy to automatize; and last but not least, we develop iii) a novel continuously acting viral selection method. The latter method is based on the assumption that antibody presentation and affinity can be used to modulate viral infection. Virus particles displaying the desired antigen can then either deliver a growth promoting or inhibiting signal. We achieve this by manipulating virus-like particles based on the recombinant Adeno-Associated Virus (rAAV) to present the antigen on the viral surface by either genetic fusion to the capsid protein or by a general enzymatic coupling step using sortase ligation technology. So far we have tested cell sorting (FACS) based on antibody expression, we have coupled YFP, which is the antigen for our test antibody system, to magentic beads, and we have produced rAAV fused at the genetic level to the antigens YFP, CFP and the sortase ligation motif. As a growth inhibiting signal our virus particles deliver a thymidine kinase gene, which enables killing by ganciclovir, and as growth promoting signal a neomycin/geneticin resistance cassette. We are in the process to determine which selection method works the best. | ||
| + | |||
| + | [[file:UP12slide7.jpg|920px|center|thumb|'''Figure 1:''' schematic illustration of the Selection Module: A virus infects cells with high affine antibodies. These cells survive. ]] | ||
=== Introduction === | === Introduction === | ||
| - | The | + | The selection module kicks in when the diverse set of antibodies is expressed on the surface of the cells. In our system each CHO cell couples the genotype of an antibody construct to the phenotype presented on the surface. Therefore, we can use all tools already available for selection of cell surface display systems. Reading literature on these methods we opted for fluorescent activated cell sorting (FACS) and magnetic beads. |
| - | |||
| - | |||
| - | |||
| - | To | + | The CHO cells express antibodies presenting them on the surface. To check if the antibodies are presented on the surface of the CHO cells we focused on different selection systems. Therefore, we used several methods like magnetic beads, preparative FACS and retargeted Adeno-associated virus (AAV). We separated the viral selection system in two parts according to the presented antibodies on the surface of the CHO cells. We designed the recombinant AAV2 to present the required antigens on the surface. One of the selection systems is based on the GFP-nanobody - YFP-antigen – complex and the other selection system is based on the anti-EGFR-antibody - EGFR complex. After a stable antigen-antibody-complex was formed, the virus infects the cells with a necessary gene for detection and selection. |
| - | + | ==== AAV2 Selection system ==== | |
| - | ===== | + | ===== Selection system of cells with Advanced Antibody Construct ===== |
| - | The | + | The first selection system is based on the expression of several fluorescence proteins. The CHO cells present on their surface the GFP-nanobody. This nanobody binds GFP and YFP with a high affinity and high specificity. The recombinant AAV carries YFP on the surface to target specifically the CHO cells with GFP-nanobody and to infect them (figure 2). |
| - | + | [[File:UP12 nonmutated-ab yfp-virus.png|600px|center|thumb|'''Figure 2:''' schematic illustration of the recombinant adeno-associated virus with YFP on the surface and CFP as gene of interest, binds to the antibody representing CHO cell]] | |
| - | + | ||
| - | |||
| - | + | To examine the effect of the enzyme AID stably transfected in the CHO cells to induce the hypersomatic mutation, we designed a recombinant virus with CFP on the surface. GFP-nanobody expressed on CHO cells binds YFP, but not the closely related CFP. Our goal was to show the AID mutates the GFP-nanobody to an antibody with a higher affinity to CFP. Formation of a stable antigen-antibody complex enables the virus binding to the cell and infect it with a mVenus-signal. By excitation the infected cells could be visualized by shining with yellow colour (figure 3). | |
| - | + | [[File:UP12 mutated-ab CFP-labled-virus.png|600px|center|thumb|'''Figure 3:''' schematic illustration of the recombinant adeno-associated virus with CFP on the surface and YFP as Gene of interest, binds to the mutate antibody representing CHO cell ]] | |
| - | |||
| - | |||
| - | ===== ''AAV2 Lytic and lysogenic pathways | + | To select the cells with the desired antigens the Virus gives the cells a survival signal by infecting them. Virus with the same CFP-antigen on the surface could not bind to the GFP-nanobody. Altered by the AID GFP-nanobody binds CFP linked to the virus following by the infection with kanamycin resistance gene cassette. The kanamycin/neomycin gene codes for the aminoglycoside phosphotransferase enzyme, which inactivates by phosphorylation the aminoglycoside antibiotics such as geneticin (G418). Cultured in the correct Geneticin concentration, which was determined by titration against non-transfected controls, selection for stable transfectants is possible (figure 4). |
| - | After entry the | + | |
| + | [[File:UP12 Xenia antibioticresistance4.png|600px|center|thumb|'''Figure 4:''' schematic illustration of the selection system using the neomycin as gene of interest and CFP on the surface binds to the mutate antibody representing CHO cell.]] | ||
| + | |||
| + | ===== Selection system of cells with Smaller Antibody Construct ===== | ||
| + | The second selection system consists of the EGFR-antigen binding to the anti-EGFR-antibody (scFv425). The scFv425 has a high affinity for EGFR-antigen. After a stable binding the cell would be transduced with transgene encoding for cyan fluorescence protein CFP. The fluorescence could detected at the CFP channel by an excitation at 405 nm and an emission at 425–475 nm. A higher transduction rate would mean the modification of anti-EGFR-antibody to a higher affinity to EGFR which was driven by Activation-induced cytidine deaminase (AID). The selection of cells with improved antibody also relies on the transduction of cells with kanamycin resistance gene cassette. | ||
| + | |||
| + | ==== AAV2 Genome ==== | ||
| + | AAV is a DNA virus with single stranded linear genome of 4.7 kb. The genome consists of two open reading frames rep and cap which are flanked by two inverted terminal repeats (ITR). | ||
| + | |||
| + | The rep gene located on the 5'-end of the genome encodes four overlapping multifunctional regulatory proteins. The transcription of the Rep proteins begins at the promoters p5 (Rep 78/Rep 68) and p19 (Rep 52/Rep 40). The two larger Rep proteins Rep78 and Rep68 are required for genome replication, the regulation of gene expression, and for site specific integration. The smaller Rep proteins Rep52 and Rep40 are involved in the encapsidation of the AAV-Genome. | ||
| + | |||
| + | The open reading frame cap on the 3'end of the AAV-genome encodes the three viral capsid proteins VP1, VP2, and VP3. The transcription of the mRNA is regulated by the p40 promoter. Alternative splicing results in two mRNA molecules: one encoding VP1 and the other VP2/ VP3. Application of an alternative splice acceptor removes the first AUG start codon for VP1. Thereby mRNA that is mainly formed encodes for the VP2 and VP3 capsid proteins. The first AUG codon in the transcript is the initiation codon for VP3. The translation of VP2 begins at an upstream ACG non-methionine start codon resulting in an approximately 10-fold lower translation of VP2. The stop codon of the three capsid proteins is identical. The stoichiometric ratio of the three capsid proteins VP1, VP2 and VP3 forming the icosahedral form is 1:1:10. | ||
| + | |||
| + | The ITRs are the indispensible cis-acting elements, which are critical for genome replication and packaging. The ITRs contain the Rep-binding site (RBS), which binds the Rep78/68 by unwinding the ITR at the terminal resolution site (trs). | ||
| + | |||
| + | ==== AAV2 Infection ==== | ||
| + | |||
| + | AAV2 viruses kill cancer cells without harming healthy ones. Viruses cross the membrane by a process called penetration. | ||
| + | |||
| + | The virus particles present in the cell media come closer to the cells by diffusion. After several attachments to the cell membrane the virus binds to the cell receptors and enter the cell by receptor mediated endocytosis. There are three cell receptors playing a major role in non-effectivity. Ubiquitously expressed cell surface Heparan sulfate proteoglycan (HSPG) functions as a primary receptor account for the broad host range of AAV. aVß5 integrin and fibroblast growth factor 1 (FGFR-1) have a co-receptor activity for successful viral entry into the host cell. Bound AAV particles enter the cell very rapidly through clathrin mediated endocytosis. Each endosome carries a single AAV particle. After internalization the virus requires acidic environment for sufficient release into the cytosol. AAV accumulates in the perinuclear space and enters through the nuclear pore complex (NPC) into the nucleus, where uncoating and gene expression take place. | ||
| + | |||
| + | ==== AAV2 Lytic and lysogenic pathways ==== | ||
| + | After entry to the nucleus, AAV follows either lytic or lysogenic life cycle. In the presence of a helper virus, Adenovirus or Herpesvirus, the AAV is directed in a lytic life cycle. AAV undergoes a productive infection, which includes rapid replication in the nucleus and the later release of the new particles into the environment by bursting cells. | ||
If there is no helper virus, AAV establish latency by integration into the host genome. The Rep expression is limited. When AAV infects a human cell, it integrates in a site-specific manner into the chromosome 19, called AAVS1. Essential components are the ITRs and the AAVS1 in cis and Rep proteins 78/68 in trans. Rep78/68 binds to the Rep-binding site (RBS) on ITR and unwinds the ITR at the terminal resolution site (trs). | If there is no helper virus, AAV establish latency by integration into the host genome. The Rep expression is limited. When AAV infects a human cell, it integrates in a site-specific manner into the chromosome 19, called AAVS1. Essential components are the ITRs and the AAVS1 in cis and Rep proteins 78/68 in trans. Rep78/68 binds to the Rep-binding site (RBS) on ITR and unwinds the ITR at the terminal resolution site (trs). | ||
| - | The productive cycle is initiated by infecting with the helper virus, what affects stronger Rep expression and episomal AAV DNA replication. Rep proteins excise the AAV genes out of the host cell’s chromosome. The excised genes are replicated in the | + | The productive cycle is initiated by infecting with the helper virus, what affects stronger Rep expression and episomal AAV DNA replication. Rep proteins excise the AAV genes out of the host cell’s chromosome. The excised genes are replicated in the nucleus and the capsid proteins package the replicated genes in the cell. When a large number of these particles have assembled, the helper virus induces cell lysis and release the newly assembled particles into the environment. |
| - | ==== | + | ==== Recombinant AAV2 ==== |
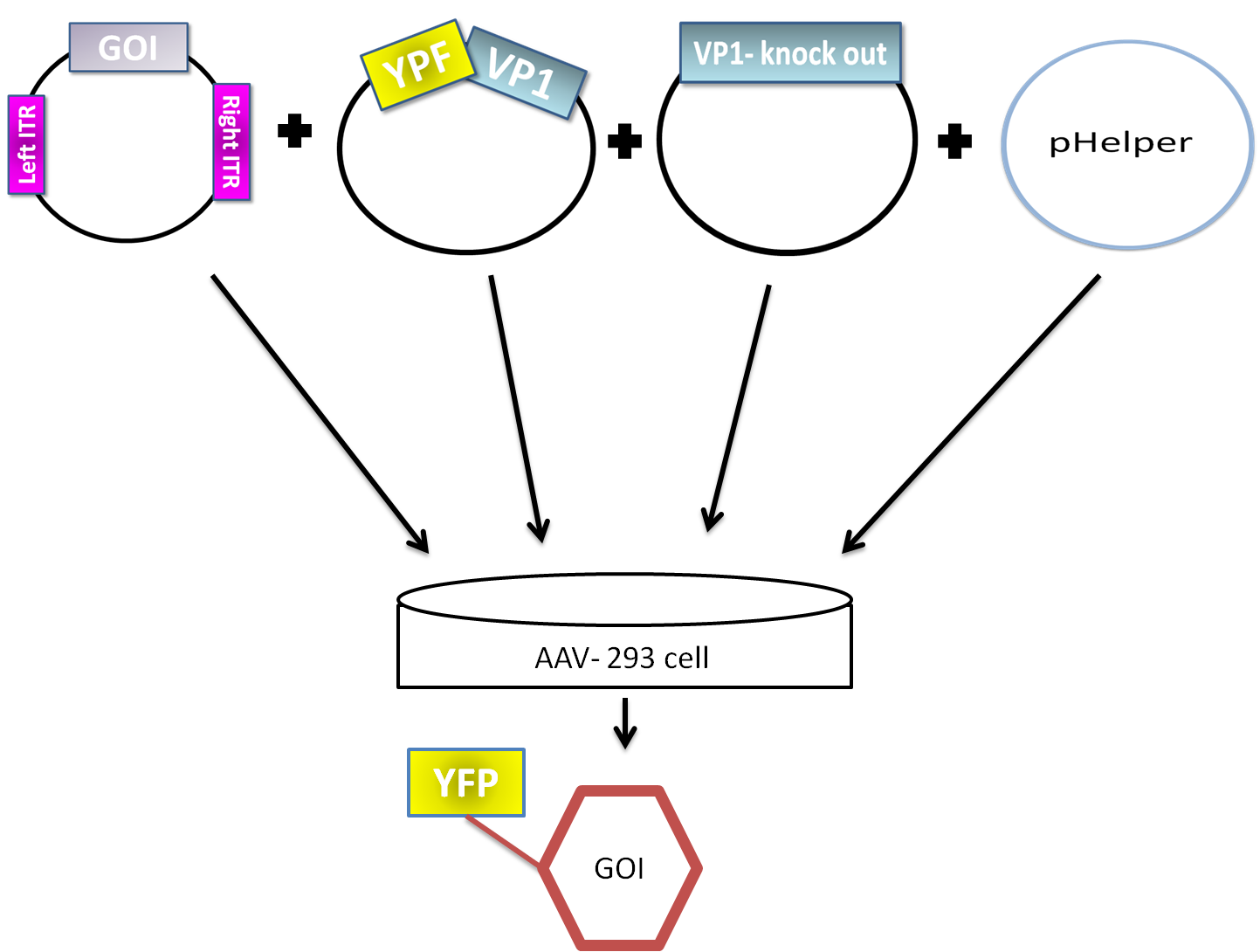
| - | AAV are replication-deficient parvoviruses | + | AAV are replication-deficient parvoviruses. Although AAV can replicate in the absence of a helper virus, they require a co-infection of adenovirus for an efficient replication. To bypass the co-infection with adenoviruses, which is associated with disadvantages like the contamination of AAV preparation, all of the required genes of adenovirus were encoded on a helperplasmid. The helperplasmid consisting of adenovirus E2A, VA und E4 genes, is transfected in the AAV-293-cells, which stably express the adenovirus E1 gene. The production of infectious adeno-associated virus particles is able by co-transfecting the AAV-293 cells with four plasmids: the plasmid that carries the helper genes taken from adenovirus, a plasmid coding the protein fused on VP1 or VP2 cap-gene, a vector plasmid that contains the wildtype AAV2 Rep-Cap genes with VP2 or VP1 knock out and an AAV2 ITR-providing plasmid carrying the gene of interest (figure 5). The transgene cassette flanked by the ITRs is wrapped as a single strand in the virus capsid. The replication of the plasmid with transgene cassette is implemented with rep-proteins and enveloped in the viral capsid formed by cap-proteins. |
| + | |||
| + | [[File:Phelper prinzip.png|600px|center|thumb|'''Figure 5:''' schematic illustration of the pHelper system for virus production in AAV-293 host cells]] | ||
| + | |||
| + | ==== EGFR-D3 ==== | ||
| + | Epidermal growth factor receptor (EGFR), also known as HER1 or ErbB1, is a member of a family of tyrosine kinase receptors which modulates tumor cell differentiation, survival, adhesion, migration and proliferation. EGFR drives tumorigenesis in many cancers such as head and neck cancer, pancreatic cancer, squamous cell lung carcinoma , and colorectal cancer. EGFR consists of three major functional domains: a cytoplasmic tyrosine kinase domain, a hydrophobic transmembrane domain, and an extracellular ligand-binding domain. | ||
| + | |||
| + | In our project we expressed only the extracellular ligand-binding domain also called 3rd domain, wich binds to the anti EGFR antibody. | ||
| + | |||
| + | ==== Sortase ==== | ||
| + | To bring big proteins like EGFR on the surface of the virus, the ligation of two polypeptides in a chemoselective manner could be done. The completely expressed protein e.g. EGFR should contain the C-terminal Sortase motif to be recognized by the enzyme Sortase. N-terminal Sortase motif was fused to the N-terminus of VP2/3 gene to allow the ligation by the Sortase. | ||
| + | |||
| + | Sortase is an enzyme which catalyzes specific ligation of two proteins to each other. In the first step the enzym recognizes the C-terminal conserved LPxTG sortase motif and cleaves this motif between Gly and Thr. The obtained thioester intermediat reacts with an N-terminal glycine, regenerating a native amide bond. | ||
| + | |||
| + | [[File:UP12 Sortase-schematic representation.jpg|400px|center|thumb|'''Figure 6:''' schematic illustration of enzymatic reaction of the sortase]] | ||
| + | |||
| + | Gram positive bacteria use the sortase to covalently ligate the proteins to the peptidoglycan layers. | ||
| + | |||
| + | ==== Magnetic beads selection system ==== | ||
| + | |||
| + | Our advanced construct presents an anti-GFP activity. Therefore, we could design a selection system that uses magnetic beads coupled to Green Fluorescence Protein (GFP) as an alternative method to select cells presenting antibody on their surface. The system is based on the creation of sequence of proteins connected to the magnetic beads.<br> | ||
| + | The magnetic beads that we used are the commercial product of Invitrogen® and they are originally coupled to streptavidin - Dynabeads®. Biotin binds to streptavidin with very high affinity and specificity. GFP is covalently coupled to the biotin. Because our antibody construct shows anti-GFP activity, it binds to GFP and can be pulled together with biotinylated GFP connected to magnetic beads with streptavidin (figure 7).<br> | ||
| + | <br> | ||
| + | [[File:UP12_Magnet.png|500px|center|thumb|'''Figure 7:''' schematic illustration of the dynabeads connected to the biotinylated GFP coupled to the CHO cell presenting antibody on the surface]] | ||
| + | <br> | ||
| + | |||
| + | ==== FACS selection system ==== | ||
| + | Fluorescent activated cell sorting enables the the characterization of each cell based on forward scatter, side scatter (i.e. size and granularity) and fluorescence in a variety of channels depending on the instrument. As at our institute the only cell sorter had only a 488 and 635 nm laser excitation, which was not suited to sort our GFP, YFP and mCHerry reporters, we turned to the cell sorting facility of the Deutsches Rheuma-Forschungszentrum Berlin (DRFZ). At this facility we tested sorting of the small antibody construct (YFP) in combination with AID expression (GFP) and the advanced antibody construct in combination (mCherry) with AID (GFP) based on the fluorescence of the reporter genes. For both constructs 300'000 double positive cells were sorted. Since the sorting was successful, we are confident that we can also sort cells based on the mCherry reporter and a fluorescently labeled antigen.<br> | ||
| + | [[File:UP12 FACSprocess.jpg|400 px|center|thumb|'''Figure 8:''' schematic illustration of Fluorescent activated cell sorting ]] | ||
=== '''Results''' === | === '''Results''' === | ||
| - | ==== ''...'' ==== | + | ==== Targeting the virus against the small antibody construct ==== |
| + | |||
| + | To target the virus against the small construct of the antibody we designed plasmids required for viral assembly. EGF-receptor (EGFR) is a big protein which would disturb the viral assembly, so we focused on the enzyme sortase which covalently attaches proteins to each other. Expressed the third EGFR domain should contain the C-terminal Sortase motif to be recognized by the enzyme Sortase. N-terminal Sortase motif was fused to the N-terminus of VP2/3 gene of adeno-associated virus in the BioBricks (BBa_K929200 and BBa_K929201) to allow the ligation of third EGFR domain on the surface of virus by the Sortase. | ||
| + | |||
| + | The first plasmid with the N-terminal sortase motif was designed. The BioBrick pCMV_DARPin-E01_Middle-Linker_[AAV2]-VP23 was used to get the fusion parts linked to the VP2/3 cap-gene. The fusion parts Kozak sequence and the N-terminal sortase motif were constructed as a forward primer close to the VP2/3 – gene, which was used as an annealing part. The reversed primer was designed to enclose the whole VP2/3 region. For the following digestion the restriction sites for XbaI and PstI were also presented in the primers. The second plasmid was designed as the first one, but it contained an additional myc-tag to recognize that the sortase motif is on the surface of the virus. | ||
| + | |||
| + | Amplificate product generated during the PCR was digested with the enzymes XbaI and PstI. Then it was ligated into the BioBrick containing the CMV promoter (figure 9)[http://partsregistry.org/Part:BBa_I712004 (BBa_I712004)]. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:UP12_Sortase_without_myc.png|600px|center|thumb|'''Figure 9:''' schematic illustration of the sortase motiv cloned into PSB1C backbone (BBa_K929202)]] | ||
| + | |||
| + | The next extracellular ligand-binding domain of EGFR was constructed to be expressed in ''E. coli''. The coding part for the ligand-binding domain was inserted downstream to the PelB sequence and upstream to the His-tag and C-terminal sortase motif. The constructed plasmid contain the RFC25 according suffix and prefix. | ||
| + | |||
| + | |||
| + | |||
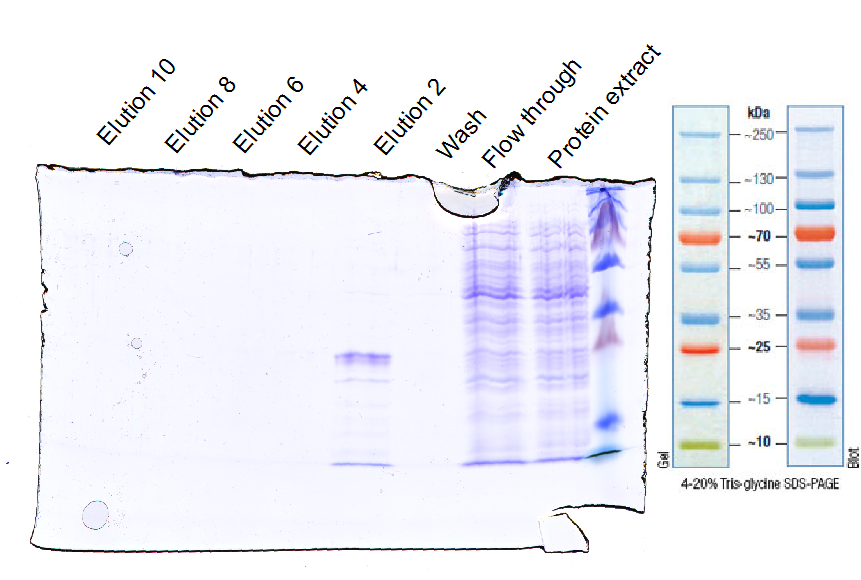
| + | To express the EGFR ligand-binding domain in XL1-Blue ''E.coli'' strain the constructed part was ligated in the arabinose inducible promoter containing plasmid (pBAD). The expression was induced with arabinose and the incubation conditions were 4 h at 30 °C (figure 10). EGFR was purified with Ni-NTA (figure 11) and concentrated for ligation with sortase. | ||
| + | <table><tr><td> | ||
| + | [[File:UP12_mainEGFR.PNG|300px|thumb|'''Figure 10:''' schematic illustration of the expression of the 3rd EGFR domain in ''E. Coli'' detection was performed using SDS-PAGE and Western-blot]] | ||
| + | </td><td> | ||
| + | [[File:UP12_SDA-PAGE-2012-10-20.png|500px|thumb|'''Figure 11:''' SDS-PAGE of purified fractions.]] | ||
| + | </td></tr></table> | ||
| + | The EGFR constructed part was also cloned into the BioBrick containing the CMV promoter. [http://partsregistry.org/Part:BBa_I712004 (Bba_I712004)]. The resulting Biobrick (BBa_K929202) corresponds to the original RFC10 prefix and suffix. But the desired fusion parts PelB, EGF-Receptor_3rd domain, His-tag and C-terminal sortase motif have 2 additional enclosing restriction sites compatible with the RFC25. That means, that the Part 'PelB, EGF-Receptor_3rd domain, His-tag and C-terminal sortase motif' could be cut out with the enzymes XbaI and AgeI (figure 11). | ||
| + | |||
| + | [[Image:UP12 BBa K929202 design3.png|600px|center|thumb|'''Figure 11:''' schematic illustration of the sortase motiv and His-tag cloned into PSB1C backbone (Bba_K929200)]] | ||
| + | |||
| + | ==== Targeting the virus against the advanced antibody construct ==== | ||
| + | |||
| + | |||
| + | To modify the surface exposed proteins of the recombinant virus the desired genes were fused to the N-terminus of VP2 cap-gene. VP2 cap-gene is not essential for viral effectivity, what makes it an ideal candidate for the insertions of sequences to retarget the particle. Therefore, to reduce and to define the AAV tropism, the primary receptor motif, heparan sulphate proteoglycan motif, was knocked-out. The plasmids YFP fused to VP1, CFP fused to VP2, mVenus and CFP as gene of interest and the plasmids with VP2 and VP1 knock out. To get the completely assembled recombinant adeno-associated virus with the YFP on the surface and CFP as gene of interest, we transfected the AAV-293 cells with four plasmids: the plasmid YFP fused to VP1, the rep-cap plasmid with VP1 knock-out, phelper plasmid and the CFP as gene of interest. | ||
| + | |||
| + | The yellow fluorescence was detectible (figure 12), pointing out that the virus was completely assembled. To check whether the viral assembly worked we infected the AAV-HT1080 cells with the recombinant virus (figure 13), like it was described in the AAV Helper-Free System Instruction Manual. The next day several HT1080 cells were gleaming cyan by the excitation. | ||
| + | |||
| + | |||
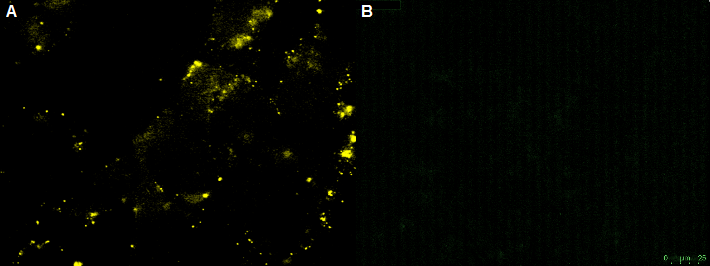
| + | [[File:UP12 CHO bleaching.PNG|center|500px|thumb|'''Figure 12:''' Fluorescence image of the negative control - non transfeced CHO-cells with the recombinant adeno-associated virus rAAV) .The rAAV are not able to infect the CHO-cells (without nanobody). A) unbleached B) bleached]] | ||
| + | |||
| + | |||
| + | <gallery widths=250px heights=250px perrow=3> | ||
| + | |||
| + | File:UP12 HT1080 infected.PNG|'''Figure 13:''' Fluorescence image of the positive control – infected HT1080 cells with the recombinant adeno-associated virus with YFP on the surface and CFP as the Gene of interest. Cyan fluorescence are the indicator for a successful infection | ||
| + | File:UP12kleinerHT1080 Sortase 3 merge.jpg|'''Figure 14:''' Fluorescence image of the positive control – infected HT1080 cells with the recombinant adeno-associated virus with Sortasemotif on the surface and CFP as the Gene of interest. Cyan fluorescence is the indicator for a successful infection | ||
| + | </gallery> | ||
| + | |||
| + | |||
| + | <br><br><br><br> | ||
| + | |||
| + | The quantitative real-time polymerase chain reaction (qPCR) is one of the leading methods of the biological research. The significant difference between PCR and qPCR is the detection ''via'' fluorescence. The change in the fluorescence intensity can be detected in each cycle. This fluorescence is proportional to the amount of the PCR product. This technique allows the user to follow the PCR reaction in real time. Because of the digital acquisition of the fluorescence, there are no more post-PCR steps (like gel electrophoresis) necessary. (Kubista, M et al. 2006)(Heid, Chr. et al. 1996). Using a standard curve of known sequence copies, the amount of DNA of the samples can be exactly detected. For this detection an intercalating fluorescent dye, like SYBR green, is used. SYBR green is an unspecific florescence dye, which binds within the minor groove of the double stranded DNA. By binding into the DNA, the fluorescent signal increases significantly and can be detected. <br> | ||
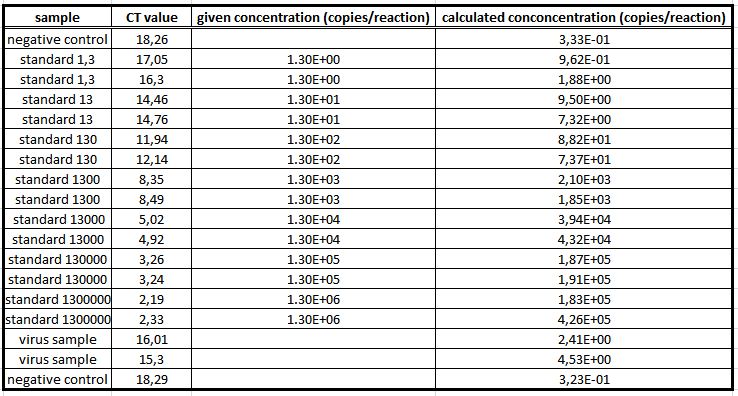
| + | We used the qPCR to determine the genomic virus titer (figure 15). Therefore, the cellular and plasmid DNA left in our virus stocks have to be digested. For that reason 5 µL of the supernatant from pelleted cell lysate was treated with 7.5 µL DNaseI and 5 µL MgCl2 (c= 50 mM) in a final volume of 50 µL. Additionally, the samples were incubated at 37 °C for 30 min with a subsequent inactivation at 65 °C for 10 min. PCR reactions were carried out with 5 µL of our digested samples. (Rohr ''et al.'' 2002, Rohr ''et al.'' 2005)(figure 14). | ||
| + | [[File: UP12_20120925qpcr.jpg|left|430px|thumb|'''Figure 14:''' qPCR: genomic virus titer - quantification curves of the qPCR]][[File: UP12_20120925cp.jpg|right|430px|thumb|'''Figure 15:''' qPCR: genomic virus titer – cp-values]]<br> | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | We also created a virus with CFP on its surface and mVenus as the gene of interest. We used the plasmids that were the vector plasmid with CFP fused to VP2, the rep-cap plasmid with VP2 knock-out, phelper plasmid and mVenus as gene of interest. The further task will be to test this virus construct by infecting the HT1080 cells. We expect yellow fluorescence of the HT1080 cells. | ||
| + | |||
| + | ==== Selection system with antibiotic resistance ==== | ||
| - | + | We designed primer to get the neomycin/kanamycin cassette into the plasmid containing the left and right ITR. To select the cells with the improved affinity of the antibody to the antigen the virus provide the cell with a survival signal. In the near feature we want to infect the cells with the virus containing the neomycin/kanamycin resistance gene cassette. The infected cells should be cultivated on media containing Gentamycin. | |
| - | + | ==== Coupling of nanobody with Magnetic Beads ==== | |
| - | + | The switch from antibody presentation to antibody secretion is induced by the Cre Recombinase. The supernatant was collected and incubated with magnetic beads coupled to GFP via biotin-streptavidin. While performing a western blot we detected a band at 41kDa which correlates with the size of the nanobody-Fc construct, showing that the expected GFP-nanobody binding occurs (figure 16). | |
| + | <br> | ||
| + | [[file:UP12_Cre_Recombinase_Blot_ohneBildunterschrift.png|center|300px|thumb|'''Figure 16:''' Western Blot of purified Nanobody-Fc from supernatant of cells co-transfected with Cre recombinase and advanced construct. Purification was carried out with magnetic beads. Correct band can be seen at 41kDa]]<br> | ||
| - | === | + | === Discussion === |
| - | + | The positive control cell line HT1080 was infected with the rAAV. A cyan fluorescence is the indication of a successful infection of the HT1080 cells. According to this we can conclude that the virus has been assembled correctly, because the virus was able to infect the HT1080 cells.<br> | |
| - | The | + | The non-transfected CHO cells with the GFP-nanobody-construct can not be infected by the rAAV. This was shown by transducing CHO cells with the rAAV. In this case the YFP virus particle are located around the CHO cell. No CFP could be detected into the cells. This shows that the rAAV is unable to infect the non-transfected CHO cells. By bleaching the rAAV the fluorescence intensity decreased. The bleaching of fluorescence is an indication of YFP-virus particle. <br> |
| - | + | The last part our virus requires to be targeted against the small antibody construct, so EGFR was expressed in ''E. coli''. The expressed protein could be detected using the SDS-PAGE and Western-blot. The size of the EGFR 3rd domain fused to the sortase motif is 24 kDa and this band was presented in the western-blot.<br> | |
| + | For testing the genomic virus titer the qPCR was used. Low copies were detected.<br> | ||
| - | + | === '''References''' === | |
| - | * | + | * Russell DW, Kay MA. Adeno-associated virus vectors and hematology. Blood. 1999 Aug 1;94(3):864-74. Review. [http://www.ncbi.nlm.nih.gov/pubmed/10419876 PubMed PMID: 10419876.] |
| - | * Bartlett | + | * Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000 Mar;74(6):2777-85. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC111768/ PubMed PMID: 10684294.] |
| - | * Kubista M.; Andrade, JM.; Bengtsson, M.; Forootan, A.; Jonak, J; Lind, K.; Sindelka, R; Sjoback, R.; Sjogreen, B.; Strombom, L.; Stahlberg, A.; Zoric, N.; “The real-time polymerase chain reaction”, Mol Aspects Med. 27(2-3):95-125, 2006. | + | * Kubista M.; Andrade, JM.; Bengtsson, M.; Forootan, A.; Jonak, J; Lind, K.; Sindelka, R; Sjoback, R.; Sjogreen, B.; Strombom, L.; Stahlberg, A.; Zoric, N.; “The real-time polymerase chain reaction”, Mol Aspects Med. 27(2-3):95-125, 2006. Apr-Jun;27(2-3):95-125. Epub 2006 Feb 3. Review. [http://www.ncbi.nlm.nih.gov/pubmed/16460794 PubMed PMID: 16460794.] |
| - | * Heid, | + | * Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996 Oct;6(10):986-94. [http://www.ncbi.nlm.nih.gov/pubmed?term=Heid%2C%20Chr.%3B%20Stevens%2C%20J.%3B%20Livak%2C%20K.%3B%20Williams%2C%20P.%3B%20%E2%80%9CReal%20Time%20Quantitative%20PCR%E2%80%9D%2C%20Genome%20Reserch%206%3A%20986-994%2C%201996 PubMed PMID: 8908518.] |
| - | * Rohr, U.-P. et al., 2005. Quantitative real-time PCR for titration of infectious recombinant AAV-2 particles. Journal of virological methods, 127(1), pp.40-5. [http://www.ncbi.nlm.nih.gov/pubmed/15893564 | + | * Rohr, U.-P. et al., 2005. Quantitative real-time PCR for titration of infectious recombinant AAV-2 particles. Journal of virological methods, 127(1), pp.40-5. [http://www.ncbi.nlm.nih.gov/pubmed/15893564 PubMed PMID: 1589356]. |
| - | * Rohr, U.-P. et al., 2002. Fast and reliable titration of recombinant adeno-associated virus type-2 using quantitative real-time PCR. Journal of virological methods, 106(1), pp.81-8. [http://www.ncbi.nlm.nih.gov/pubmed/12367732 | + | * Rohr, U.-P. et al., 2002. Fast and reliable titration of recombinant adeno-associated virus type-2 using quantitative real-time PCR. Journal of virological methods, 106(1), pp.81-8. [http://www.ncbi.nlm.nih.gov/pubmed/12367732 PubMed PMID: 12367732] |
| + | *Levary DA, Parthasarathy R, Boder ET, Ackerman ME. Protein-protein fusion catalyzed by sortase A. PLoS One. 2011 Apr 6;6(4):e18342. PubMed PMID: 21494692; PubMed Central [http://www.ncbi.nlm.nih.gov/pubmed?term=Protein-Protein%20Fusion%20Catalyzed%20by%20Sortase%20A PMCID: PMC3071835.] | ||
| + | *Smith RL, Traul DL, Schaack J, Clayton GH, Staley KJ, Wilcox CL. Characterization of promoter function and cell-type-specific expression from viral vectors in the nervous system. J Virol. 2000 Dec;74(23):11254-61. PubMed PMID: 11070024; PubMed Central [http://www.ncbi.nlm.nih.gov/pubmed?term=Characterization%20of%20Promoter%20Function%20and%20Cell-Type-Specific%20Expression%20from%20Viral%20Vectors%20in%20the%20Nervous%20System PMCID: PMC113226.] | ||
| + | * Ward P, Walsh CE. Targeted integration of a rAAV vector into the AAVS1 region. Virology. 2012 Sep 12. pii: S0042-6822(12)00396-0. doi: 10.1016/j.virol.2012.08.015. [Epub ahead of print] [http://www.ncbi.nlm.nih.gov/pubmed/22981435 PubMed PMID: 22981435.] | ||
| + | * | ||
Latest revision as of 17:29, 26 October 2012
Contents
|
Selection Module
Abstract Selection Module
The "selection module" comprises a set of methods to select the desired cell clones from the collection of cells each expressing one variant of the antibody construct. Depending on the needs, different selection methods might have specific advantages. We set up three methods, whereby two are well established but discontinuous: i) fluorescent activated cell sorting (FACS), which yields many data but requires expensive instrumentation and ii) magnetic beads, which are less expensive and easy to automatize; and last but not least, we develop iii) a novel continuously acting viral selection method. The latter method is based on the assumption that antibody presentation and affinity can be used to modulate viral infection. Virus particles displaying the desired antigen can then either deliver a growth promoting or inhibiting signal. We achieve this by manipulating virus-like particles based on the recombinant Adeno-Associated Virus (rAAV) to present the antigen on the viral surface by either genetic fusion to the capsid protein or by a general enzymatic coupling step using sortase ligation technology. So far we have tested cell sorting (FACS) based on antibody expression, we have coupled YFP, which is the antigen for our test antibody system, to magentic beads, and we have produced rAAV fused at the genetic level to the antigens YFP, CFP and the sortase ligation motif. As a growth inhibiting signal our virus particles deliver a thymidine kinase gene, which enables killing by ganciclovir, and as growth promoting signal a neomycin/geneticin resistance cassette. We are in the process to determine which selection method works the best.
Introduction
The selection module kicks in when the diverse set of antibodies is expressed on the surface of the cells. In our system each CHO cell couples the genotype of an antibody construct to the phenotype presented on the surface. Therefore, we can use all tools already available for selection of cell surface display systems. Reading literature on these methods we opted for fluorescent activated cell sorting (FACS) and magnetic beads.
The CHO cells express antibodies presenting them on the surface. To check if the antibodies are presented on the surface of the CHO cells we focused on different selection systems. Therefore, we used several methods like magnetic beads, preparative FACS and retargeted Adeno-associated virus (AAV). We separated the viral selection system in two parts according to the presented antibodies on the surface of the CHO cells. We designed the recombinant AAV2 to present the required antigens on the surface. One of the selection systems is based on the GFP-nanobody - YFP-antigen – complex and the other selection system is based on the anti-EGFR-antibody - EGFR complex. After a stable antigen-antibody-complex was formed, the virus infects the cells with a necessary gene for detection and selection.
AAV2 Selection system
Selection system of cells with Advanced Antibody Construct
The first selection system is based on the expression of several fluorescence proteins. The CHO cells present on their surface the GFP-nanobody. This nanobody binds GFP and YFP with a high affinity and high specificity. The recombinant AAV carries YFP on the surface to target specifically the CHO cells with GFP-nanobody and to infect them (figure 2).
To examine the effect of the enzyme AID stably transfected in the CHO cells to induce the hypersomatic mutation, we designed a recombinant virus with CFP on the surface. GFP-nanobody expressed on CHO cells binds YFP, but not the closely related CFP. Our goal was to show the AID mutates the GFP-nanobody to an antibody with a higher affinity to CFP. Formation of a stable antigen-antibody complex enables the virus binding to the cell and infect it with a mVenus-signal. By excitation the infected cells could be visualized by shining with yellow colour (figure 3).
To select the cells with the desired antigens the Virus gives the cells a survival signal by infecting them. Virus with the same CFP-antigen on the surface could not bind to the GFP-nanobody. Altered by the AID GFP-nanobody binds CFP linked to the virus following by the infection with kanamycin resistance gene cassette. The kanamycin/neomycin gene codes for the aminoglycoside phosphotransferase enzyme, which inactivates by phosphorylation the aminoglycoside antibiotics such as geneticin (G418). Cultured in the correct Geneticin concentration, which was determined by titration against non-transfected controls, selection for stable transfectants is possible (figure 4).
Selection system of cells with Smaller Antibody Construct
The second selection system consists of the EGFR-antigen binding to the anti-EGFR-antibody (scFv425). The scFv425 has a high affinity for EGFR-antigen. After a stable binding the cell would be transduced with transgene encoding for cyan fluorescence protein CFP. The fluorescence could detected at the CFP channel by an excitation at 405 nm and an emission at 425–475 nm. A higher transduction rate would mean the modification of anti-EGFR-antibody to a higher affinity to EGFR which was driven by Activation-induced cytidine deaminase (AID). The selection of cells with improved antibody also relies on the transduction of cells with kanamycin resistance gene cassette.
AAV2 Genome
AAV is a DNA virus with single stranded linear genome of 4.7 kb. The genome consists of two open reading frames rep and cap which are flanked by two inverted terminal repeats (ITR).
The rep gene located on the 5'-end of the genome encodes four overlapping multifunctional regulatory proteins. The transcription of the Rep proteins begins at the promoters p5 (Rep 78/Rep 68) and p19 (Rep 52/Rep 40). The two larger Rep proteins Rep78 and Rep68 are required for genome replication, the regulation of gene expression, and for site specific integration. The smaller Rep proteins Rep52 and Rep40 are involved in the encapsidation of the AAV-Genome.
The open reading frame cap on the 3'end of the AAV-genome encodes the three viral capsid proteins VP1, VP2, and VP3. The transcription of the mRNA is regulated by the p40 promoter. Alternative splicing results in two mRNA molecules: one encoding VP1 and the other VP2/ VP3. Application of an alternative splice acceptor removes the first AUG start codon for VP1. Thereby mRNA that is mainly formed encodes for the VP2 and VP3 capsid proteins. The first AUG codon in the transcript is the initiation codon for VP3. The translation of VP2 begins at an upstream ACG non-methionine start codon resulting in an approximately 10-fold lower translation of VP2. The stop codon of the three capsid proteins is identical. The stoichiometric ratio of the three capsid proteins VP1, VP2 and VP3 forming the icosahedral form is 1:1:10.
The ITRs are the indispensible cis-acting elements, which are critical for genome replication and packaging. The ITRs contain the Rep-binding site (RBS), which binds the Rep78/68 by unwinding the ITR at the terminal resolution site (trs).
AAV2 Infection
AAV2 viruses kill cancer cells without harming healthy ones. Viruses cross the membrane by a process called penetration.
The virus particles present in the cell media come closer to the cells by diffusion. After several attachments to the cell membrane the virus binds to the cell receptors and enter the cell by receptor mediated endocytosis. There are three cell receptors playing a major role in non-effectivity. Ubiquitously expressed cell surface Heparan sulfate proteoglycan (HSPG) functions as a primary receptor account for the broad host range of AAV. aVß5 integrin and fibroblast growth factor 1 (FGFR-1) have a co-receptor activity for successful viral entry into the host cell. Bound AAV particles enter the cell very rapidly through clathrin mediated endocytosis. Each endosome carries a single AAV particle. After internalization the virus requires acidic environment for sufficient release into the cytosol. AAV accumulates in the perinuclear space and enters through the nuclear pore complex (NPC) into the nucleus, where uncoating and gene expression take place.
AAV2 Lytic and lysogenic pathways
After entry to the nucleus, AAV follows either lytic or lysogenic life cycle. In the presence of a helper virus, Adenovirus or Herpesvirus, the AAV is directed in a lytic life cycle. AAV undergoes a productive infection, which includes rapid replication in the nucleus and the later release of the new particles into the environment by bursting cells.
If there is no helper virus, AAV establish latency by integration into the host genome. The Rep expression is limited. When AAV infects a human cell, it integrates in a site-specific manner into the chromosome 19, called AAVS1. Essential components are the ITRs and the AAVS1 in cis and Rep proteins 78/68 in trans. Rep78/68 binds to the Rep-binding site (RBS) on ITR and unwinds the ITR at the terminal resolution site (trs).
The productive cycle is initiated by infecting with the helper virus, what affects stronger Rep expression and episomal AAV DNA replication. Rep proteins excise the AAV genes out of the host cell’s chromosome. The excised genes are replicated in the nucleus and the capsid proteins package the replicated genes in the cell. When a large number of these particles have assembled, the helper virus induces cell lysis and release the newly assembled particles into the environment.
Recombinant AAV2
AAV are replication-deficient parvoviruses. Although AAV can replicate in the absence of a helper virus, they require a co-infection of adenovirus for an efficient replication. To bypass the co-infection with adenoviruses, which is associated with disadvantages like the contamination of AAV preparation, all of the required genes of adenovirus were encoded on a helperplasmid. The helperplasmid consisting of adenovirus E2A, VA und E4 genes, is transfected in the AAV-293-cells, which stably express the adenovirus E1 gene. The production of infectious adeno-associated virus particles is able by co-transfecting the AAV-293 cells with four plasmids: the plasmid that carries the helper genes taken from adenovirus, a plasmid coding the protein fused on VP1 or VP2 cap-gene, a vector plasmid that contains the wildtype AAV2 Rep-Cap genes with VP2 or VP1 knock out and an AAV2 ITR-providing plasmid carrying the gene of interest (figure 5). The transgene cassette flanked by the ITRs is wrapped as a single strand in the virus capsid. The replication of the plasmid with transgene cassette is implemented with rep-proteins and enveloped in the viral capsid formed by cap-proteins.
EGFR-D3
Epidermal growth factor receptor (EGFR), also known as HER1 or ErbB1, is a member of a family of tyrosine kinase receptors which modulates tumor cell differentiation, survival, adhesion, migration and proliferation. EGFR drives tumorigenesis in many cancers such as head and neck cancer, pancreatic cancer, squamous cell lung carcinoma , and colorectal cancer. EGFR consists of three major functional domains: a cytoplasmic tyrosine kinase domain, a hydrophobic transmembrane domain, and an extracellular ligand-binding domain.
In our project we expressed only the extracellular ligand-binding domain also called 3rd domain, wich binds to the anti EGFR antibody.
Sortase
To bring big proteins like EGFR on the surface of the virus, the ligation of two polypeptides in a chemoselective manner could be done. The completely expressed protein e.g. EGFR should contain the C-terminal Sortase motif to be recognized by the enzyme Sortase. N-terminal Sortase motif was fused to the N-terminus of VP2/3 gene to allow the ligation by the Sortase.
Sortase is an enzyme which catalyzes specific ligation of two proteins to each other. In the first step the enzym recognizes the C-terminal conserved LPxTG sortase motif and cleaves this motif between Gly and Thr. The obtained thioester intermediat reacts with an N-terminal glycine, regenerating a native amide bond.
Gram positive bacteria use the sortase to covalently ligate the proteins to the peptidoglycan layers.
Magnetic beads selection system
Our advanced construct presents an anti-GFP activity. Therefore, we could design a selection system that uses magnetic beads coupled to Green Fluorescence Protein (GFP) as an alternative method to select cells presenting antibody on their surface. The system is based on the creation of sequence of proteins connected to the magnetic beads.
The magnetic beads that we used are the commercial product of Invitrogen® and they are originally coupled to streptavidin - Dynabeads®. Biotin binds to streptavidin with very high affinity and specificity. GFP is covalently coupled to the biotin. Because our antibody construct shows anti-GFP activity, it binds to GFP and can be pulled together with biotinylated GFP connected to magnetic beads with streptavidin (figure 7).
FACS selection system
Fluorescent activated cell sorting enables the the characterization of each cell based on forward scatter, side scatter (i.e. size and granularity) and fluorescence in a variety of channels depending on the instrument. As at our institute the only cell sorter had only a 488 and 635 nm laser excitation, which was not suited to sort our GFP, YFP and mCHerry reporters, we turned to the cell sorting facility of the Deutsches Rheuma-Forschungszentrum Berlin (DRFZ). At this facility we tested sorting of the small antibody construct (YFP) in combination with AID expression (GFP) and the advanced antibody construct in combination (mCherry) with AID (GFP) based on the fluorescence of the reporter genes. For both constructs 300'000 double positive cells were sorted. Since the sorting was successful, we are confident that we can also sort cells based on the mCherry reporter and a fluorescently labeled antigen.
Results
Targeting the virus against the small antibody construct
To target the virus against the small construct of the antibody we designed plasmids required for viral assembly. EGF-receptor (EGFR) is a big protein which would disturb the viral assembly, so we focused on the enzyme sortase which covalently attaches proteins to each other. Expressed the third EGFR domain should contain the C-terminal Sortase motif to be recognized by the enzyme Sortase. N-terminal Sortase motif was fused to the N-terminus of VP2/3 gene of adeno-associated virus in the BioBricks (BBa_K929200 and BBa_K929201) to allow the ligation of third EGFR domain on the surface of virus by the Sortase.
The first plasmid with the N-terminal sortase motif was designed. The BioBrick pCMV_DARPin-E01_Middle-Linker_[AAV2]-VP23 was used to get the fusion parts linked to the VP2/3 cap-gene. The fusion parts Kozak sequence and the N-terminal sortase motif were constructed as a forward primer close to the VP2/3 – gene, which was used as an annealing part. The reversed primer was designed to enclose the whole VP2/3 region. For the following digestion the restriction sites for XbaI and PstI were also presented in the primers. The second plasmid was designed as the first one, but it contained an additional myc-tag to recognize that the sortase motif is on the surface of the virus.
Amplificate product generated during the PCR was digested with the enzymes XbaI and PstI. Then it was ligated into the BioBrick containing the CMV promoter (figure 9)[http://partsregistry.org/Part:BBa_I712004 (BBa_I712004)].
The next extracellular ligand-binding domain of EGFR was constructed to be expressed in E. coli. The coding part for the ligand-binding domain was inserted downstream to the PelB sequence and upstream to the His-tag and C-terminal sortase motif. The constructed plasmid contain the RFC25 according suffix and prefix.
To express the EGFR ligand-binding domain in XL1-Blue E.coli strain the constructed part was ligated in the arabinose inducible promoter containing plasmid (pBAD). The expression was induced with arabinose and the incubation conditions were 4 h at 30 °C (figure 10). EGFR was purified with Ni-NTA (figure 11) and concentrated for ligation with sortase.
The EGFR constructed part was also cloned into the BioBrick containing the CMV promoter. [http://partsregistry.org/Part:BBa_I712004 (Bba_I712004)]. The resulting Biobrick (BBa_K929202) corresponds to the original RFC10 prefix and suffix. But the desired fusion parts PelB, EGF-Receptor_3rd domain, His-tag and C-terminal sortase motif have 2 additional enclosing restriction sites compatible with the RFC25. That means, that the Part 'PelB, EGF-Receptor_3rd domain, His-tag and C-terminal sortase motif' could be cut out with the enzymes XbaI and AgeI (figure 11).
Targeting the virus against the advanced antibody construct
To modify the surface exposed proteins of the recombinant virus the desired genes were fused to the N-terminus of VP2 cap-gene. VP2 cap-gene is not essential for viral effectivity, what makes it an ideal candidate for the insertions of sequences to retarget the particle. Therefore, to reduce and to define the AAV tropism, the primary receptor motif, heparan sulphate proteoglycan motif, was knocked-out. The plasmids YFP fused to VP1, CFP fused to VP2, mVenus and CFP as gene of interest and the plasmids with VP2 and VP1 knock out. To get the completely assembled recombinant adeno-associated virus with the YFP on the surface and CFP as gene of interest, we transfected the AAV-293 cells with four plasmids: the plasmid YFP fused to VP1, the rep-cap plasmid with VP1 knock-out, phelper plasmid and the CFP as gene of interest.
The yellow fluorescence was detectible (figure 12), pointing out that the virus was completely assembled. To check whether the viral assembly worked we infected the AAV-HT1080 cells with the recombinant virus (figure 13), like it was described in the AAV Helper-Free System Instruction Manual. The next day several HT1080 cells were gleaming cyan by the excitation.
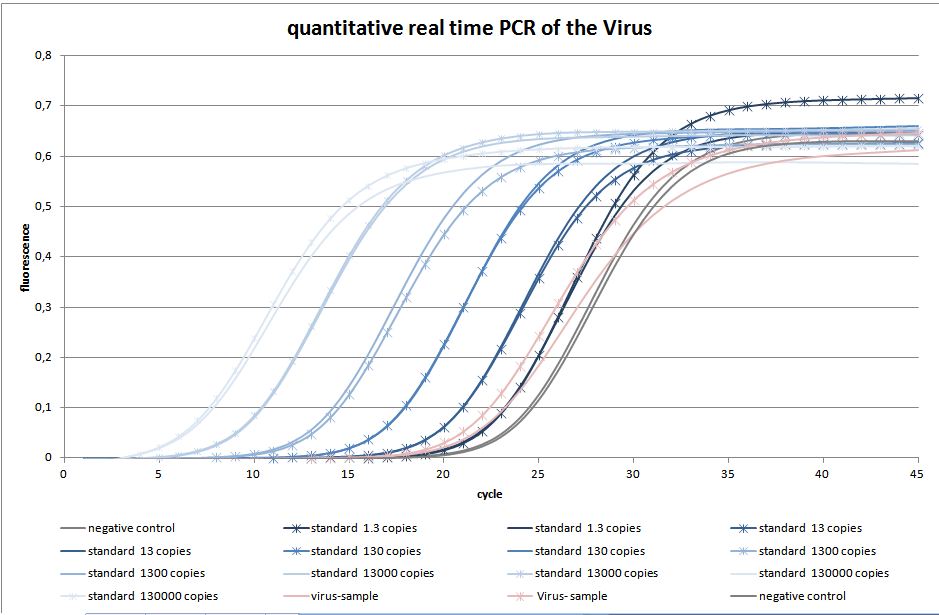
The quantitative real-time polymerase chain reaction (qPCR) is one of the leading methods of the biological research. The significant difference between PCR and qPCR is the detection via fluorescence. The change in the fluorescence intensity can be detected in each cycle. This fluorescence is proportional to the amount of the PCR product. This technique allows the user to follow the PCR reaction in real time. Because of the digital acquisition of the fluorescence, there are no more post-PCR steps (like gel electrophoresis) necessary. (Kubista, M et al. 2006)(Heid, Chr. et al. 1996). Using a standard curve of known sequence copies, the amount of DNA of the samples can be exactly detected. For this detection an intercalating fluorescent dye, like SYBR green, is used. SYBR green is an unspecific florescence dye, which binds within the minor groove of the double stranded DNA. By binding into the DNA, the fluorescent signal increases significantly and can be detected.
We used the qPCR to determine the genomic virus titer (figure 15). Therefore, the cellular and plasmid DNA left in our virus stocks have to be digested. For that reason 5 µL of the supernatant from pelleted cell lysate was treated with 7.5 µL DNaseI and 5 µL MgCl2 (c= 50 mM) in a final volume of 50 µL. Additionally, the samples were incubated at 37 °C for 30 min with a subsequent inactivation at 65 °C for 10 min. PCR reactions were carried out with 5 µL of our digested samples. (Rohr et al. 2002, Rohr et al. 2005)(figure 14).
We also created a virus with CFP on its surface and mVenus as the gene of interest. We used the plasmids that were the vector plasmid with CFP fused to VP2, the rep-cap plasmid with VP2 knock-out, phelper plasmid and mVenus as gene of interest. The further task will be to test this virus construct by infecting the HT1080 cells. We expect yellow fluorescence of the HT1080 cells.
Selection system with antibiotic resistance
We designed primer to get the neomycin/kanamycin cassette into the plasmid containing the left and right ITR. To select the cells with the improved affinity of the antibody to the antigen the virus provide the cell with a survival signal. In the near feature we want to infect the cells with the virus containing the neomycin/kanamycin resistance gene cassette. The infected cells should be cultivated on media containing Gentamycin.
Coupling of nanobody with Magnetic Beads
The switch from antibody presentation to antibody secretion is induced by the Cre Recombinase. The supernatant was collected and incubated with magnetic beads coupled to GFP via biotin-streptavidin. While performing a western blot we detected a band at 41kDa which correlates with the size of the nanobody-Fc construct, showing that the expected GFP-nanobody binding occurs (figure 16).
Discussion
The positive control cell line HT1080 was infected with the rAAV. A cyan fluorescence is the indication of a successful infection of the HT1080 cells. According to this we can conclude that the virus has been assembled correctly, because the virus was able to infect the HT1080 cells.
The non-transfected CHO cells with the GFP-nanobody-construct can not be infected by the rAAV. This was shown by transducing CHO cells with the rAAV. In this case the YFP virus particle are located around the CHO cell. No CFP could be detected into the cells. This shows that the rAAV is unable to infect the non-transfected CHO cells. By bleaching the rAAV the fluorescence intensity decreased. The bleaching of fluorescence is an indication of YFP-virus particle.
The last part our virus requires to be targeted against the small antibody construct, so EGFR was expressed in E. coli. The expressed protein could be detected using the SDS-PAGE and Western-blot. The size of the EGFR 3rd domain fused to the sortase motif is 24 kDa and this band was presented in the western-blot.
For testing the genomic virus titer the qPCR was used. Low copies were detected.
References
- Russell DW, Kay MA. Adeno-associated virus vectors and hematology. Blood. 1999 Aug 1;94(3):864-74. Review. [http://www.ncbi.nlm.nih.gov/pubmed/10419876 PubMed PMID: 10419876.]
- Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000 Mar;74(6):2777-85. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC111768/ PubMed PMID: 10684294.]
- Kubista M.; Andrade, JM.; Bengtsson, M.; Forootan, A.; Jonak, J; Lind, K.; Sindelka, R; Sjoback, R.; Sjogreen, B.; Strombom, L.; Stahlberg, A.; Zoric, N.; “The real-time polymerase chain reaction”, Mol Aspects Med. 27(2-3):95-125, 2006. Apr-Jun;27(2-3):95-125. Epub 2006 Feb 3. Review. [http://www.ncbi.nlm.nih.gov/pubmed/16460794 PubMed PMID: 16460794.]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996 Oct;6(10):986-94. [http://www.ncbi.nlm.nih.gov/pubmed?term=Heid%2C%20Chr.%3B%20Stevens%2C%20J.%3B%20Livak%2C%20K.%3B%20Williams%2C%20P.%3B%20%E2%80%9CReal%20Time%20Quantitative%20PCR%E2%80%9D%2C%20Genome%20Reserch%206%3A%20986-994%2C%201996 PubMed PMID: 8908518.]
- Rohr, U.-P. et al., 2005. Quantitative real-time PCR for titration of infectious recombinant AAV-2 particles. Journal of virological methods, 127(1), pp.40-5. [http://www.ncbi.nlm.nih.gov/pubmed/15893564 PubMed PMID: 1589356].
- Rohr, U.-P. et al., 2002. Fast and reliable titration of recombinant adeno-associated virus type-2 using quantitative real-time PCR. Journal of virological methods, 106(1), pp.81-8. [http://www.ncbi.nlm.nih.gov/pubmed/12367732 PubMed PMID: 12367732]
- Levary DA, Parthasarathy R, Boder ET, Ackerman ME. Protein-protein fusion catalyzed by sortase A. PLoS One. 2011 Apr 6;6(4):e18342. PubMed PMID: 21494692; PubMed Central [http://www.ncbi.nlm.nih.gov/pubmed?term=Protein-Protein%20Fusion%20Catalyzed%20by%20Sortase%20A PMCID: PMC3071835.]
- Smith RL, Traul DL, Schaack J, Clayton GH, Staley KJ, Wilcox CL. Characterization of promoter function and cell-type-specific expression from viral vectors in the nervous system. J Virol. 2000 Dec;74(23):11254-61. PubMed PMID: 11070024; PubMed Central [http://www.ncbi.nlm.nih.gov/pubmed?term=Characterization%20of%20Promoter%20Function%20and%20Cell-Type-Specific%20Expression%20from%20Viral%20Vectors%20in%20the%20Nervous%20System PMCID: PMC113226.]
- Ward P, Walsh CE. Targeted integration of a rAAV vector into the AAVS1 region. Virology. 2012 Sep 12. pii: S0042-6822(12)00396-0. doi: 10.1016/j.virol.2012.08.015. [Epub ahead of print] [http://www.ncbi.nlm.nih.gov/pubmed/22981435 PubMed PMID: 22981435.]
 "
"