Team:Kyoto/MethodAndMaterial
From 2012.igem.org
(→Miniprep) |
(→DNS reaegnt) |
||
| (75 intermediate revisions not shown) | |||
| Line 18: | Line 18: | ||
<ul class="kyoto-post-it"> | <ul class="kyoto-post-it"> | ||
<li>[[FIle:Kyoto_General.png|30px|link=#General]]</li> | <li>[[FIle:Kyoto_General.png|30px|link=#General]]</li> | ||
| - | <li>[[File:Kyoto_FlowerFairyEcoli_postit.png|30px|link=# | + | <li>[[File:Kyoto_FlowerFairyEcoli_postit.png|30px|link=#Flower Fairy E.coli]]</li> |
<li>[[File:Kyoto_GGA_postit.png|30px|link=#Golden Gate Assembly]]</li> | <li>[[File:Kyoto_GGA_postit.png|30px|link=#Golden Gate Assembly]]</li> | ||
</ul> | </ul> | ||
| - | = | + | =[[File:Kyoto_Protocol.png|link=]]= |
==General== | ==General== | ||
==PCR== | ==PCR== | ||
| Line 67: | Line 67: | ||
|} | |} | ||
*Let stand for 2min at 94℃.<br> | *Let stand for 2min at 94℃.<br> | ||
| - | *25-40 cycles for 10s at 98℃, for 30s at Tm-5℃, and for 1min (1min for 1kb) at 68℃ (Tm is | + | *25-40 cycles for 10s at 98℃, for 30s at Tm-5℃, and for 1min (1min for 1kb) at 68℃ (Tm is temperature at which primer will dissolve).<br> |
*Agarose Gel Electrophoresis for confirmation.<br><br> | *Agarose Gel Electrophoresis for confirmation.<br><br> | ||
'''PCR: Takara Ex taq'''<br> | '''PCR: Takara Ex taq'''<br> | ||
| Line 87: | Line 87: | ||
|} | |} | ||
*Dispense 25µL to 15 tubes.<br> | *Dispense 25µL to 15 tubes.<br> | ||
| - | *Pick a single colony and transfer it to each | + | *Pick a single colony and transfer it to each tube.<br> |
*Suspend the colony.<br> | *Suspend the colony.<br> | ||
*Let stand for 10min at 90℃.<br> | *Let stand for 10min at 90℃.<br> | ||
| Line 95: | Line 95: | ||
*Agalose Gel Electrophoresis for confirmation.<br> | *Agalose Gel Electrophoresis for confirmation.<br> | ||
*Negative Control: Use nothing.<br> | *Negative Control: Use nothing.<br> | ||
| - | *Positive Control: Use a colony that will yield a product with | + | *Positive Control: Use a colony that will yield a product with these primers.<br> |
==RNA Extraction== | ==RNA Extraction== | ||
| Line 115: | Line 115: | ||
==Making Competent cells== | ==Making Competent cells== | ||
| - | *Streak E.coli cells on an LB plate; (BL21(DE3)LysS cells on LB plate+34 mg/ml chloramphenicol)<br> | + | *Streak ''E.coli'' cells on an LB plate; (BL21(DE3)LysS cells on LB plate+34 mg/ml chloramphenicol)<br> |
| - | *Allow cells to grow at | + | *Allow cells to grow at 37℃ overnight<br> |
| - | *Place one colony in 10 mL LB media (+antibiotic selection if necessary), grow overnight at | + | *Place one colony in 10 mL LB media (+antibiotic selection if necessary), grow overnight at 37℃<br> |
*Take 2 ml LB media and save for blank. Transfer 5 mL overnight DH5a culture into 500 mL LB media in 3 L flask<br> | *Take 2 ml LB media and save for blank. Transfer 5 mL overnight DH5a culture into 500 mL LB media in 3 L flask<br> | ||
| - | *Allow cell to grow at | + | *Allow cell to grow at 37℃ (250 rpm), until OD600= 0.4 (~2-3 hours)<br> |
*Transfer cells to 2 centrifuge bottles (250 mL), and place cells on ice for 20 mins<br> | *Transfer cells to 2 centrifuge bottles (250 mL), and place cells on ice for 20 mins<br> | ||
| - | *Centrifuge cells in Sorval GSA rotor at | + | *Centrifuge cells in Sorval GSA rotor at 4℃ for 10 mins at 3,000 g.<br> |
*Subsequent resuspensions may be done in the same bottle. Cells must remain cold for the rest of the procedure: Transport tubes on ice and resuspend on ice in the cold room<br> | *Subsequent resuspensions may be done in the same bottle. Cells must remain cold for the rest of the procedure: Transport tubes on ice and resuspend on ice in the cold room<br> | ||
*Pour off media and resuspend cells in 30 mL of cold 0.1 M CaCl2. Transfer the suspended cells into 50 mL polypropylene falcon tubes, and incubate on ice for 30 mins<br> | *Pour off media and resuspend cells in 30 mL of cold 0.1 M CaCl2. Transfer the suspended cells into 50 mL polypropylene falcon tubes, and incubate on ice for 30 mins<br> | ||
| - | *Centrifuge cells using Sorval RT6000B rotor at | + | *Centrifuge cells using Sorval RT6000B rotor at 4℃ for 10 mins at 3,000 g (2500 rpm)<br> |
| - | *Pour supernatant and resuspend cells (by pipetting) in 8 mL cold 0.1M CaCl2 containing 15% glycerol. Transfer 140 mL into (1.5 mL)<br> *Ependorff tubes placed on ice. Freeze the cells in liquid nitrogen. Cells stored at - | + | *Pour supernatant and resuspend cells (by pipetting) in 8 mL cold 0.1M CaCl2 containing 15% glycerol. Transfer 140 mL into (1.5 mL)<br> |
| + | *Ependorff tubes placed on ice. Freeze the cells in liquid nitrogen. Cells stored at -80℃ can be used for transformation for up to ~6 months<br> | ||
| + | |||
==Miniprep== | ==Miniprep== | ||
Use QIAprep Spin Miniprep Kit Cat. No. 27104 by QIAGEN<br> | Use QIAprep Spin Miniprep Kit Cat. No. 27104 by QIAGEN<br> | ||
| Line 132: | Line 134: | ||
*Transfer a half of the culture to a tube.<br> | *Transfer a half of the culture to a tube.<br> | ||
*Harvest the bacterial cells by centrifugation at 14,000g for 1min at 4℃. Remove the medium by decanting.<br> | *Harvest the bacterial cells by centrifugation at 14,000g for 1min at 4℃. Remove the medium by decanting.<br> | ||
| - | *Transfer the half of the culture to same tube and harvest as same. Remove the medium by | + | *Transfer the half of the culture to same tube and harvest as same. Remove the medium by pipetting.<br> |
| - | *Resuspend pelleted bacterial cells in 250µL Buffer P1 and mix thoroughly by | + | *Resuspend pelleted bacterial cells in 250µL Buffer P1 and mix thoroughly by pipeting.<br> |
*Add 250µL Buffer P2 and mix thoroughly by inverting the tube gently 4-6 times.<br> | *Add 250µL Buffer P2 and mix thoroughly by inverting the tube gently 4-6 times.<br> | ||
*Add 350µL Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.<br> | *Add 350µL Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.<br> | ||
| Line 140: | Line 142: | ||
*Centrifuge for 10s in a table-top microcentrifuge. Discard the flow-through.<br> | *Centrifuge for 10s in a table-top microcentrifuge. Discard the flow-through.<br> | ||
*Wash the QIAprep spin column by adding 0.5mL Buffer PB and centrifuging for 10s in a table-top microcentrifuge. Discard the flow-through.<br> | *Wash the QIAprep spin column by adding 0.5mL Buffer PB and centrifuging for 10s in a table-top microcentrifuge. Discard the flow-through.<br> | ||
| - | *Wash QIAprep spin column by adding 0.65mL Buffer PeE and centrifuging | + | *Wash QIAprep spin column by adding 0.65mL Buffer PeE and centrifuging for 10s in a table-top microcentrifuge.<br> |
*Discard the flow-through, and centrifuge for and additional 1min to remove residual wash buffer.<br> | *Discard the flow-through, and centrifuge for and additional 1min to remove residual wash buffer.<br> | ||
*Place the QIAprep column in a clean tube. To elute DNA, add 50µL water to the center of each QIAprep spin column, let stand for 1min, and centrifuge for 1min.<br> | *Place the QIAprep column in a clean tube. To elute DNA, add 50µL water to the center of each QIAprep spin column, let stand for 1min, and centrifuge for 1min.<br> | ||
| Line 148: | Line 150: | ||
*Agarose Gel Electrophoresis for Confirmation.<br> | *Agarose Gel Electrophoresis for Confirmation.<br> | ||
| - | Use | + | Use Wizard Plus SV Minipreps DNA Purification System by Promega |
*Harvest 1–5ml (high-copy-number plasmid) or 10ml (low-copy-number plasmid) of bacterial culture by centrifugation for 5 minutes at 10,000 x g in a tabletop centrifuge. Pour off the supernatant and blot the inverted tube on a paper towel to remove excess media.<br> | *Harvest 1–5ml (high-copy-number plasmid) or 10ml (low-copy-number plasmid) of bacterial culture by centrifugation for 5 minutes at 10,000 x g in a tabletop centrifuge. Pour off the supernatant and blot the inverted tube on a paper towel to remove excess media.<br> | ||
*Add 250µl of Cell Resuspension Solution and completely resuspend the cell pellet by vortexing or pipetting. It is essential to thoroughly resuspend the cells. If they are not already in a microcentrifuge tube, transfer the resuspended cells to a sterile 1.5ml microcentrifuge tube(s).<br> | *Add 250µl of Cell Resuspension Solution and completely resuspend the cell pellet by vortexing or pipetting. It is essential to thoroughly resuspend the cells. If they are not already in a microcentrifuge tube, transfer the resuspended cells to a sterile 1.5ml microcentrifuge tube(s).<br> | ||
| - | *Add 250µl of Cell Lysis Solution and mix by inverting the tube 4 times | + | *Add 250µl of Cell Lysis Solution and mix by inverting the tube 4 times (do not vortex). Incubate until the cell suspension clears (approximately 1–5 minutes).<br> |
| - | (do not vortex). Incubate until the cell suspension clears (approximately 1–5 minutes).<br> | + | |
*Add 10µl of Alkaline Protease Solution and mix by inverting the tube 4 times. Incubate for 5 minutes at room temperature.<br> | *Add 10µl of Alkaline Protease Solution and mix by inverting the tube 4 times. Incubate for 5 minutes at room temperature.<br> | ||
*Add 350µl of Neutralization Solution and immediately mix by inverting the tube 4 times (do not vortex).<br> | *Add 350µl of Neutralization Solution and immediately mix by inverting the tube 4 times (do not vortex).<br> | ||
| Line 179: | Line 180: | ||
*Precipitation.<br> | *Precipitation.<br> | ||
| - | == | + | ==Electrophoresis== |
*Prepare 200mL of a 1.0% agarose solution:<br> | *Prepare 200mL of a 1.0% agarose solution:<br> | ||
*Measure 2.0g agarose into a beaker.<br> | *Measure 2.0g agarose into a beaker.<br> | ||
| Line 196: | Line 197: | ||
*Place the tray in electrophoresis chamber.<br> | *Place the tray in electrophoresis chamber.<br> | ||
*Cover the tray with 1xTAE buffer.<br> | *Cover the tray with 1xTAE buffer.<br> | ||
| - | *To prepare samples for | + | *To prepare samples for electrophoresis, add 1µL of 6x Loading Buffer for every 5µL of DNA solution and mix well.<br> |
*Load 6µL of the DNA solution per well.<br> | *Load 6µL of the DNA solution per well.<br> | ||
| - | * | + | *Electrophoresis at 100V for about 30min until Loading Buffer have migrated approximately three-quarters of the gel.<br> |
*Stain the gel in 0.5µg/mL ethidium bromide for 20-30min.<br> | *Stain the gel in 0.5µg/mL ethidium bromide for 20-30min.<br> | ||
*Rinse the gel with MilliQ.<br> | *Rinse the gel with MilliQ.<br> | ||
| Line 207: | Line 208: | ||
*Turn off the transilluminator.<br> | *Turn off the transilluminator.<br> | ||
*Dispose of the gel.<br> | *Dispose of the gel.<br> | ||
| + | |||
==PCR Purification== | ==PCR Purification== | ||
Use Wizard SV Gel and PCR Clean-Up System by Promega.<br> | Use Wizard SV Gel and PCR Clean-Up System by Promega.<br> | ||
| Line 235: | Line 237: | ||
|total||36μL | |total||36μL | ||
|} | |} | ||
| - | *Mix the reaction mix thoroughly ,and dispense 36μL into 96 wells plate.<br> | + | *Mix the reaction mix thoroughly, and dispense 36μL into 96 wells plate.<br> |
*Add primer set 9μL.<br> | *Add primer set 9μL.<br> | ||
*Mix by inverting and voltex.<br> | *Mix by inverting and voltex.<br> | ||
| Line 242: | Line 244: | ||
*40 cycles for 15sec at 95°C, for 30sec at 60°C, and for 1min at 72°C.<br> | *40 cycles for 15sec at 95°C, for 30sec at 60°C, and for 1min at 72°C.<br> | ||
*Let stand for 15sec at 95°C.<br> | *Let stand for 15sec at 95°C.<br> | ||
| + | |||
==Ligation== | ==Ligation== | ||
*Make 2µL of Mixture (the vector and the insert at 1 : 5-10) and Control (only the vector).<br> | *Make 2µL of Mixture (the vector and the insert at 1 : 5-10) and Control (only the vector).<br> | ||
*Add 5µL Ligation High, 1µL T4 Kinase, and 7µL MilliQ to create a solution.<br> | *Add 5µL Ligation High, 1µL T4 Kinase, and 7µL MilliQ to create a solution.<br> | ||
| - | *Incubate at 16℃ for 30 min. If the colonies of E.coli transformed with the Control<br> | + | *Incubate at 16℃ for 30 min. If the colonies of ''E.coli'' transformed with the Control<br> |
| + | |||
==Restrictive Digestion== | ==Restrictive Digestion== | ||
Use EcoRI, XbaI, SpeI, PstI, (NEB)<br> | Use EcoRI, XbaI, SpeI, PstI, (NEB)<br> | ||
| Line 262: | Line 266: | ||
'''M9 medium'''<br> | '''M9 medium'''<br> | ||
*Stir Na2HPO4 6 g, KH2PO4 3 g, NaCl 0.5 g and NH4Cl 1 g with water 1L.<br> | *Stir Na2HPO4 6 g, KH2PO4 3 g, NaCl 0.5 g and NH4Cl 1 g with water 1L.<br> | ||
| - | *After autoclave, add 10 ml filter sterilized 100 mM MgSO4, 20 % glucose, 10 mM CaCl2, 100 mM thiamine-HCl.<br> | + | *After autoclave, add 10 ml filter sterilized 100 mM MgSO4, 20 % glucose, 10 mM CaCl2, and 100 mM thiamine-HCl.<br> |
If you need, add 10 ml filter sterilized 20 % casamino acid.<br> | If you need, add 10 ml filter sterilized 20 % casamino acid.<br> | ||
'''LB medium'''<br> | '''LB medium'''<br> | ||
| Line 282: | Line 286: | ||
*Add water up to 200 ml.<br> | *Add water up to 200 ml.<br> | ||
*Filter sterilize.<br> | *Filter sterilize.<br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Solubilization of Antibiotics== | ==Solubilization of Antibiotics== | ||
| Line 311: | Line 307: | ||
*Unfreeze conpitent cells on ice.<br> | *Unfreeze conpitent cells on ice.<br> | ||
*Dry a plate by letting the plate upside down and partly open in incubator.<br> | *Dry a plate by letting the plate upside down and partly open in incubator.<br> | ||
| - | *Add 1µL DNA solution and 20µL | + | *Add 1µL DNA solution and 20µL competent cells to 1.5mL tube, let stand for 30min on ice. If few colonies are observed, increase the amount of the competent cells or DNA, but make the amount of DNA not to get over that of the competent cells.<br> |
*Heatshock for 60s at 42℃.<br> | *Heatshock for 60s at 42℃.<br> | ||
*Let stand for 2min on ice.<br> | *Let stand for 2min on ice.<br> | ||
| - | *Culture for 1h in preculture medium (LB or SOC medium), and plate by using spreader. Do not heat spreader too much because | + | *Culture for 1h in preculture medium (LB or SOC medium), and plate by using spreader. Do not heat spreader too much because ''E.coli'' will dead for heat.<br> |
==Sequence== | ==Sequence== | ||
| Line 334: | Line 330: | ||
*Add 25µL 100% ethanol and 1µL NAOAC<br> | *Add 25µL 100% ethanol and 1µL NAOAC<br> | ||
| - | == | + | ==Flower Fairy E.coli== |
===Western blotting=== | ===Western blotting=== | ||
=====Gel solution===== | =====Gel solution===== | ||
| Line 443: | Line 439: | ||
#Add secondary antibody conjugated with AP and incubate with shaking for 30 min. at RT.<br> | #Add secondary antibody conjugated with AP and incubate with shaking for 30 min. at RT.<br> | ||
#Wash with TBST and incubate with shaking for 10 min, three times.<br> | #Wash with TBST and incubate with shaking for 10 min, three times.<br> | ||
| - | #Add 66µL NBT and 33µLof BCIP into color development buffer | + | #Add 66µL NBT and 33µLof BCIP into color development buffer, and add the mixture to the membrane.<br> |
#After the coloring, wash with DW. | #After the coloring, wash with DW. | ||
| Line 460: | Line 456: | ||
Method:<br> | Method:<br> | ||
| - | 1. Peel cuticles on parafilm by using the head of pencil.(Menasha,wI,54952)<br> | + | 1. Peel cuticles on parafilm by using the head of pencil. (Menasha, wI, 54952)<br> |
2. Put plant cells into GFP&R9 or GFP for 5~30min.<br> | 2. Put plant cells into GFP&R9 or GFP for 5~30min.<br> | ||
3. Put plant cells into PBS.<br> | 3. Put plant cells into PBS.<br> | ||
| Line 476: | Line 472: | ||
===Cultivation of ''Arabidopsis thaliana''=== | ===Cultivation of ''Arabidopsis thaliana''=== | ||
| - | '''Preparation''' | + | '''Preparation''' |
*MS culture medium | *MS culture medium | ||
*mixed salts for MS culture medium | *mixed salts for MS culture medium | ||
| Line 491: | Line 487: | ||
|} | |} | ||
*sucrose | *sucrose | ||
| - | *0 | + | *0.1N KOH |
*Gellan gum or Gelrite | *Gellan gum or Gelrite | ||
*sterilized solution (Prepare this solution when you need it.) | *sterilized solution (Prepare this solution when you need it.) | ||
| Line 503: | Line 499: | ||
|} | |} | ||
*sterilized water | *sterilized water | ||
| - | *sterilized by heating 0 | + | *sterilized by heating 0.1%(w/v) agar |
*70% ethanol | *70% ethanol | ||
'''Condition'''<br> | '''Condition'''<br> | ||
| Line 520: | Line 516: | ||
*Centrifuge a microtube lightly, and dump supernatant. | *Centrifuge a microtube lightly, and dump supernatant. | ||
*Repeat steps of [6. to 7.] 3~5 times. | *Repeat steps of [6. to 7.] 3~5 times. | ||
| - | *Suspend seeds in a few hundreds~ one thousand μl of 0 | + | *Suspend seeds in a few hundreds~ one thousand μl of 0.1% agar solution. |
*Take proper quantity of seeds with a micropipette, and drop it on culture medium. | *Take proper quantity of seeds with a micropipette, and drop it on culture medium. | ||
| + | ===Extruction of protein in E.coli with lysozyme=== | ||
| + | *Harvest the bacterial cells by centrifugation at 3,000g for 15min at 4℃. | ||
| + | *Resuspend pelleted bacterial cells in the quantity of 1/20 of the culture field of lysis buffer. | ||
| + | *Add 1/50 of the whole of lysozyme solution. | ||
| + | *Mix gently for 30min on ice. | ||
| + | *Add 1/50~1/10 of the whole of 10% Triton X-100. | ||
| + | *Mix for 20 min at 4℃. | ||
| + | *Remove cell debris and unneeded things by centrifugation at 12,000g for 30min at 4℃. | ||
| + | *Collect the clear top of the liquid. | ||
| + | notes<br> | ||
| + | Lysis buffer : 50 mM Tris-HCl or Phosphate, 150 mM NaCl, 1 mM PMSF, pH8.0<br> | ||
| + | Lysozyme solution : 10 mg/ml lysozyme in 20 mM Tris-HCl, pH8.0<br><br> | ||
| + | ===Purification of protein with His Tag=== | ||
| + | *Put 1/10~1/20 of lysate of Ni-NTA resin on column. | ||
| + | *Wash Ni-NTA resin with 3 times of whole amount of column of PBS. | ||
| + | *Equilibrate Ni-NTA resin with 3 times of whole amount of column of Equilibration buffer. | ||
| + | **(Add equilibration buffer to column and cover under nozzle of column with palafilm then leave for 30min in order to drop equilibration buffer) | ||
| + | *Bond proteins with Ni-NTA resin adding lysate to column with equilibrated Ni-NTA resin | ||
| + | *Wash with 10 times of the whole of wash buffer | ||
| + | *Elution with 5 times of the whole of elution buffer | ||
| + | *Collect Fraction by an equal amount | ||
| + | notes<br> | ||
| + | Equilibration buffer : 20mM Tris-HCl(pH7.5) 150mM NaCl<br> | ||
| + | Wash buffer : Equilibration buffer + 20mM imidazole<br> | ||
| + | Elution buffer : Equilibration buffer + 200mM imidazole<br> | ||
| + | |||
| + | ==Confocal microscope== | ||
| + | '''Making a prepared specimen''' | ||
| + | *PFA 500μL | ||
| + | *MilliQ 2100μL | ||
| + | *Hoechst 1μL | ||
| + | *Slide glass | ||
| + | #Gather fungus body by centrifugal separation.<br> | ||
| + | #Add 500μL of the PFA to a microtube containing fungus body and resuspend pellet.<br> | ||
| + | #Keep shading it for 10minutes at 4℃.<br> | ||
| + | #Centrifuge it for 1minute at 16,000×g and discard the supernatant.<br> | ||
| + | #Add 1000μL of MilliQ and 1μL of Hoechst to the microtube, resuspend pellet and incubate bacterial suspension for 30minutes or more.<br> | ||
| + | #Centrifuge bacterial suspension for 1minute at 16,000×g and discard the supernatant.<br> | ||
| + | #Add 1000μL of MilliQ, resuspend pellet, centrifuge bacterial suspension and discard the supernatant again.<br> | ||
| + | #Add 100μL of MilliQ and resuspend pellet.<br> | ||
| + | #Pipette bacterial suspension on a slide glass and dry it.<br> | ||
| + | #Preserve it with shading it.<br> | ||
| + | |||
| + | =Golden Gate Assembly= | ||
| + | ===Preparation for “Golden Gate Assembly=== | ||
| + | *Design of primers for PCR | ||
| + | There are some rules in designing primers for PCR, when you make primers to get PCR productions for “Golden Gate Assembly”. Firstly you have to design them as you can add an adapter, which includes BsaI recognition site and BsaI restriction site, to PCR productions. (The BsaI recognition sequence "GGTCTC" is separated from its four bp overhang by a single bp, and BsaI activity is independent of the sequences of the single bp spacer and the four bp overhang.) Secondly you need to design primers as any two parts in a bin are completely interchangeable with respect to Golden Gate assembly, and only a single pair of oligos is required for each part across the entire assembly.<br><br> | ||
| + | |||
| + | ===Golden Gate Assembly Protocol=== | ||
| + | *Add 100 ng of the linearized vector backbone and equimolar amounts of the other assembly pieces to a 15 µl total volume assembly reaction mixture as follows: | ||
| + | |||
| + | {|class="wikitable" | ||
| + | !linearized vector backbone||each additional assembly piece||10x NEB T4 Buffer||100x BSA||BsaI||NEB T4 Ligase, 2 million cohesive end units / mL||dH2O | ||
| + | |- | ||
| + | |100 ng||to equimolar with backbone||1.5 µl||0.15 µl||1µl||1 µl||to 15µl | ||
| + | |} | ||
| + | |||
| + | :NOTE: It is essential to use a High Concentration Ligase | ||
| + | :BsaI is only 10% active at 37 C without the addition of BSA. | ||
| + | |||
| + | *Perform the assembly reaction in a thermocycler as follows: | ||
| + | either (following Engler 2009):<br> | ||
| + | 3 min @ 37 C }<br> | ||
| + | 4 min @ 16 C } 25 cycles<br> | ||
| + | 5 min @ 50 C }<br> | ||
| + | 5 min @ 80 C } 1 cycle<br> | ||
| + | *Transform 5 µl of the assembly reaction into 100 µl of competent ''E. coli'' and/or run a diagnostic agarose gel to check for successful assembly. | ||
</div> | </div> | ||
<div id="kyoto-tab-parts"> | <div id="kyoto-tab-parts"> | ||
| + | |||
| + | We constructed these BioBrick parts in 2012!! | ||
| + | |||
<groupparts>iGEM012 Kyoto</groupparts> | <groupparts>iGEM012 Kyoto</groupparts> | ||
| + | |||
| + | This year, we define FT protein [[Part:BBa_K797006 |'''(BBa_K797006) ''']], FT generator with His-tag [[Part:BBa_K797015 |'''(BBa_K797015) ''']], T7-6His-R9 [[Part:BBa_ K797014 |'''(BBa_ K797014) ''']], RBS-TorA [[Part:BBa_K797002 |'''(BBa_K797002) ''']], and Plasmid Backbone for golden gate assembly [[Part:BBa_ K797013 |'''(BBa_ K797013) ''']] as our favorite BioBrick parts of 2012.We confirmed that these parts function properly.<br><br> | ||
| + | |||
| + | |||
| + | ===FT protein [[Part:BBa_K797006 |(BBa_K797006)]]=== | ||
| + | [[File:FT.png|thumb|200px|left|Construction of FT]] | ||
| + | FLOWERING LOCUS T(FT) protein, is an identity of florigen, plants' hormone that induce flowering at a low dose. This protein is the transcription factor for promoting the expression of other genes, such as API and FUL.<br><br> | ||
| + | |||
| + | === FT generator with His-tag [[Part:BBa_K797015 |(BBa_K797015)]]=== | ||
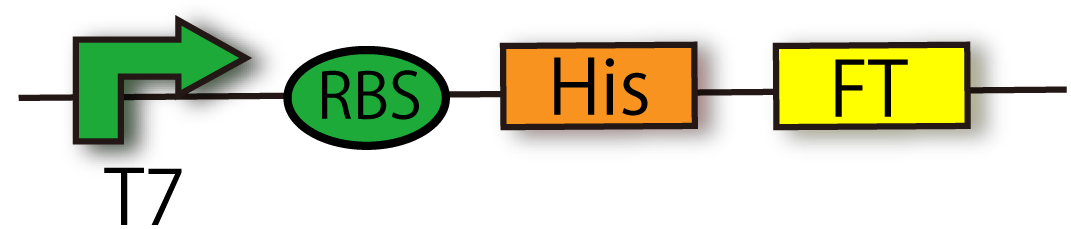
| + | [[File:T7RBSHisFT.png|thumb|300px|right|Construction of T7-RBS-His-FT]] | ||
| + | This device is composed of T7 promoter [[Part:BBa_I712074 |'''(BBa_I712074)''']], RBS and FT gene [[Part:BBa_K797006|'''(BBa_K797006)''']] with Histag. This device doesn't have terminator so that you can combine any part you like, such as GFP, with FT generator. | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
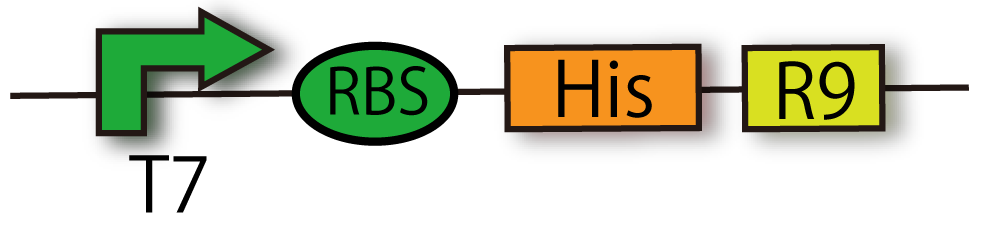
| + | === T7-RBS-6His-R9 [[Part:BBa_ K797014 |(BBa_ K797014)]]=== | ||
| + | [[File:T7RBSHisR9.png|thumb|300px|left|Construction of T7-RBS-His-R9]] | ||
| + | We can extract proteins by His tag, and penetrate proteins into cells by R9 sequence. | ||
| + | <br><br><br><br><br> | ||
| + | |||
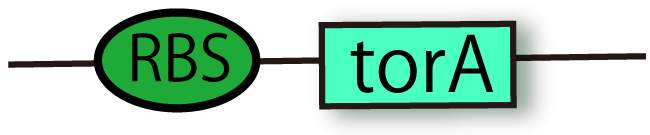
| + | === RBS-TorA [[Part:BBa_K797002 |(BBa_K797002)]]=== | ||
| + | [[File:RBStorA.png|thumb|250px|right|Construction of RBS-TorA]] | ||
| + | This part is coding N-terminal TorA signal region with RBS. Proteins conjugated TorA signal are recognized by Tat secretion pathway and they are secreted from cytoplasm to periplasm while maintaining their holdings. This comes from the part with the enzyme of TMAO reductase. This part includes RBS so that you don't bother to insert new RBS between this signal and your promoter. | ||
| + | |||
| + | ===Plasmid Backbone for golden gate assembly[[Part:BBa_ K797013 |(BBa_ K797013)]]=== | ||
| + | This backbone is suitable for golden gate assembly. Golden gate assembly is a suitable assembly to combine many parts in order you want. This plasmid backbone has restriction enzyme cleavage site of Dpn1, let you build construction by Golden gate assembly in one ligation. | ||
| + | |||
| + | We conducted Golden gate assembly by ourselves and confirmed this part has the restriction cite and restriction enzyme cutting site of Bsa1. Please see detail on Experiment page of this part. | ||
| + | |||
| + | |||
| + | |||
</div> | </div> | ||
<div id="kyoto-tab-material"> | <div id="kyoto-tab-material"> | ||
==Strains== | ==Strains== | ||
| - | ===E.coli=== | + | ===''E.coli''=== |
'''JM109''' endA, recA1, gyrA96,thi,hsdR17,(rκ-,mκ+),relA1, supE44,λ-,Δ(lac-proAB),[F',traD36,proAB,laclqZΔM15] <br> | '''JM109''' endA, recA1, gyrA96,thi,hsdR17,(rκ-,mκ+),relA1, supE44,λ-,Δ(lac-proAB),[F',traD36,proAB,laclqZΔM15] <br> | ||
'''BL21(DE3)''' F-, ompT, hsdSB(rB -mB - ), gal, dcm, (DE3)<br> | '''BL21(DE3)''' F-, ompT, hsdSB(rB -mB - ), gal, dcm, (DE3)<br> | ||
===Plant=== | ===Plant=== | ||
| - | Arabidopsis thaliana ecotype Columbia | + | ''Arabidopsis thaliana'' ecotype Columbia |
==Primers== | ==Primers== | ||
====Florigen==== | ====Florigen==== | ||
| Line 577: | Line 673: | ||
|- | |- | ||
|AP1 reverse||CTTGGTTCTGCTGATCCCACA | |AP1 reverse||CTTGGTTCTGCTGATCCCACA | ||
| + | |- | ||
| + | |pspA forward||ggaattcgcggccgcttctagagCTGGCATGTTGCTGTTGATTC | ||
| + | |- | ||
| + | |pspA reverse||aactgcagcggccgctactagtaATGCAGCCATAACCAGATCG | ||
| + | |- | ||
| + | |TatABCD forward||ggaattcgcggccgcttctagagGTCTGATCGCCTGGTTTGTC | ||
| + | |- | ||
| + | |TatABCD reverse||aactgcagcggccgctactagtaCAGTGTGAAGAATACCGAGTTCC | ||
| + | |- | ||
| + | |TMAO reductase forward||ggaattcgcggccgcttctagagTAAGGCTCACGGCGATAAGA | ||
| + | |- | ||
| + | |TMAO reductase reverse||aactgcagcggccgctactagtaCGATTTGCGTCAGTTGTTCA | ||
| + | |- | ||
| + | |kil forward||ggaattcgcggccgcttctagagTCCAAATGGGATTGCTAGGAC | ||
| + | |- | ||
| + | |kil reverse||aactgcagcggccgctactagtaGTAGCTCTTGATCCGGCAAAC | ||
| + | |- | ||
| + | | | ||
| + | |- | ||
| + | | | ||
| + | |for sequence | ||
| + | |- | ||
| + | |TatABCD forward||tcaggtgtgggcatttatcg | ||
| + | |- | ||
| + | |TatABCD reverse||gaccacaaagtaggcgaatgc | ||
| + | |- | ||
| + | |TMAOreductase forward||cgtggatgattgtggttctg | ||
| + | |- | ||
| + | |TMAOreductase reverse||gtgctgctgtagcctttgtagt | ||
| + | |- | ||
|} | |} | ||
Latest revision as of 15:56, 25 October 2012
General
PCR
PCR: ToYoBo KOD FX or ToYoBo KOD PLUS
- Dilute template DNA. If the concentration of DNA is 2-100ng/µL, transfer 1µL to a clean tube and add 99µL MilliQ.
- Dilute Primer. If the concentration of Primer is XµM, dilute primer X timers and transfer 1µL to a clean tube and add 99µL MilliQ.
- Mix the following.
For use of KOD plus ver2
| 25mM MgSO4 | 3µL |
| 2mM dNTPs | 5µL |
| 10xBuffer for KOD plus ver.2 | 5µL |
| Template DNA (5ng/µL) | 5µL |
| Primer Forward (10µM) | 1.5µL |
| Primer Reverse (10µM) | 1.5µL |
| KOD plus ver.2 | 1µL |
| MilliQ | 28µL |
| Total | 50µL |
For use of KOD FX
| 2mM dNTPs | 10µL |
| 2xBuffer for KOD FX | 25µL |
| Template DNA | 5µL |
| Primer Forward (10µM) | 1.5µL |
| Primer Reverse (10µM) | 1.5µL |
| KOD FX | 1µL |
| MilliQ | 6µL |
| Total | 50µL |
- Let stand for 2min at 94℃.
- 25-40 cycles for 10s at 98℃, for 30s at Tm-5℃, and for 1min (1min for 1kb) at 68℃ (Tm is temperature at which primer will dissolve).
- Agarose Gel Electrophoresis for confirmation.
PCR: Takara Ex taq
- Mix the following (Do on PCR Bench).
| 10x PCR buffer (TAKARA) | 40µL |
| 2.5mM dNTP | 8µL |
| Primer-1 (10pmol/µL) | 8µL |
| Primer-2 (10pmol/µL) | 8µL |
| Ex Taq HS (TAKARA) | 1.6µL |
| MilliQ | 334µL (to total 400µL) |
| Total | 400µL |
- Dispense 25µL to 15 tubes.
- Pick a single colony and transfer it to each tube.
- Suspend the colony.
- Let stand for 10min at 90℃.
- 35 cycles for 30s at 94℃, for 30s 55℃, and for 1min at 72℃.
- Let stand for 4min at 72℃.
- Add 5mL Loading Buffer to the tubes.
- Agalose Gel Electrophoresis for confirmation.
- Negative Control: Use nothing.
- Positive Control: Use a colony that will yield a product with these primers.
RNA Extraction
Use ISOGEN-LS(NIPPON GENE,311-02621
- Add 1mL ISOGEN-LS to sample and vortex.
- Store for 5min on ice.
- Add 250µL chloroform and shake vigorously for 15 sec.
- Store for 3min. on ice.
- Centrifuge 17400xg for 10min. at 4℃.
- Transfer aqueous phase to another tube and add 0.8 volume isopropanol.
- Store for 10min. on ice.
- Centrifuge 17400xg for 10min. at 4℃.
- Discard the supernatant.
- Add 800µL 80% ethanol and vortex.
- Centrifuge 7500xg for 5min. at 4℃.
- Discard the supernatant.
- Dry briefly.
- Dissolve in nuclease-free water.
Making Competent cells
- Streak E.coli cells on an LB plate; (BL21(DE3)LysS cells on LB plate+34 mg/ml chloramphenicol)
- Allow cells to grow at 37℃ overnight
- Place one colony in 10 mL LB media (+antibiotic selection if necessary), grow overnight at 37℃
- Take 2 ml LB media and save for blank. Transfer 5 mL overnight DH5a culture into 500 mL LB media in 3 L flask
- Allow cell to grow at 37℃ (250 rpm), until OD600= 0.4 (~2-3 hours)
- Transfer cells to 2 centrifuge bottles (250 mL), and place cells on ice for 20 mins
- Centrifuge cells in Sorval GSA rotor at 4℃ for 10 mins at 3,000 g.
- Subsequent resuspensions may be done in the same bottle. Cells must remain cold for the rest of the procedure: Transport tubes on ice and resuspend on ice in the cold room
- Pour off media and resuspend cells in 30 mL of cold 0.1 M CaCl2. Transfer the suspended cells into 50 mL polypropylene falcon tubes, and incubate on ice for 30 mins
- Centrifuge cells using Sorval RT6000B rotor at 4℃ for 10 mins at 3,000 g (2500 rpm)
- Pour supernatant and resuspend cells (by pipetting) in 8 mL cold 0.1M CaCl2 containing 15% glycerol. Transfer 140 mL into (1.5 mL)
- Ependorff tubes placed on ice. Freeze the cells in liquid nitrogen. Cells stored at -80℃ can be used for transformation for up to ~6 months
Miniprep
Use QIAprep Spin Miniprep Kit Cat. No. 27104 by QIAGEN
- Pick a single colony from a freshly streaked selective plate and inoculate a culture of about 3mL LB medium containing the appropriate selective antibiotic.
- Incubate at 170rpm for 8h at 37℃ with vigorous shaking.
- Transfer a half of the culture to a tube.
- Harvest the bacterial cells by centrifugation at 14,000g for 1min at 4℃. Remove the medium by decanting.
- Transfer the half of the culture to same tube and harvest as same. Remove the medium by pipetting.
- Resuspend pelleted bacterial cells in 250µL Buffer P1 and mix thoroughly by pipeting.
- Add 250µL Buffer P2 and mix thoroughly by inverting the tube gently 4-6 times.
- Add 350µL Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
- Centrifuge for 10min at 14,000g at 4℃.
- Apply the supernatants from step 10 to the QIAprep spin column by pipetting.
- Centrifuge for 10s in a table-top microcentrifuge. Discard the flow-through.
- Wash the QIAprep spin column by adding 0.5mL Buffer PB and centrifuging for 10s in a table-top microcentrifuge. Discard the flow-through.
- Wash QIAprep spin column by adding 0.65mL Buffer PeE and centrifuging for 10s in a table-top microcentrifuge.
- Discard the flow-through, and centrifuge for and additional 1min to remove residual wash buffer.
- Place the QIAprep column in a clean tube. To elute DNA, add 50µL water to the center of each QIAprep spin column, let stand for 1min, and centrifuge for 1min.
- Discard the QIAprep spin column.
- Measure the concentration of DNA by using eppendorf BioPhotometer plus.
- Restriction Digestion.
- Agarose Gel Electrophoresis for Confirmation.
Use Wizard Plus SV Minipreps DNA Purification System by Promega
- Harvest 1–5ml (high-copy-number plasmid) or 10ml (low-copy-number plasmid) of bacterial culture by centrifugation for 5 minutes at 10,000 x g in a tabletop centrifuge. Pour off the supernatant and blot the inverted tube on a paper towel to remove excess media.
- Add 250µl of Cell Resuspension Solution and completely resuspend the cell pellet by vortexing or pipetting. It is essential to thoroughly resuspend the cells. If they are not already in a microcentrifuge tube, transfer the resuspended cells to a sterile 1.5ml microcentrifuge tube(s).
- Add 250µl of Cell Lysis Solution and mix by inverting the tube 4 times (do not vortex). Incubate until the cell suspension clears (approximately 1–5 minutes).
- Add 10µl of Alkaline Protease Solution and mix by inverting the tube 4 times. Incubate for 5 minutes at room temperature.
- Add 350µl of Neutralization Solution and immediately mix by inverting the tube 4 times (do not vortex).
- Centrifuge the bacterial lysate at maximum speed (around 14,000 × g) in a microcentrifuge for 10 minutes at room temperature.
- Transfer the cleared lysate (approximately 850µl, Section 3.B, Step 6) to the prepared Spin Column by decanting. Avoid disturbing or transferring any of the white precipitate with the supernatant.
- Centrifuge the supernatant at maximum speed in a microcentrifuge for 1 minute at room temperature. Remove the Spin Column from the tube and discard the flowthrough from the Collection Tube. Reinsert the Spin Column into the Collection Tube.
- Add 750µl of Column Wash Solution, previously diluted with 95% ethanol, to the Spin Column.
- Centrifuge at maximum speed in a microcentrifuge for 1 minute at room temperature. Remove the Spin Column from the tube and discard the flowthrough. Reinsert the Spin Column into the Collection Tube.
- Repeat the wash procedure using 250µl of Column Wash Solution.
- Centrifuge at maximum speed in a microcentrifuge for 2 minutes at room temperature.om temperature.
- Transfer the Spin Column to a new, sterile 1.5ml microcentrifuge tube, being careful not to transfer any of the Column Wash Solution with the Spin Column. If the Spin Column has Column Wash Solution associated with it, centrifuge again for 1 minute at maximum speed.
- Transfer the Spin Column to a new, sterile 1.5ml microcentrifuge tube.
- Elute the plasmid DNA by adding 100µl of Nuclease-Free Water to the Spin Column. Centrifuge at maximum speed for 1 minute at room temperature in microcentrifuge.
- After eluting the DNA, remove the assembly from the 1.5ml microcentrifuge tube and discard the Spin Column.
- DNA is stable in water without addition of a buffer if stored at –20°C or below. DNA is stable at 4°C in TE buffer. To store the DNA in TE buffer, add 11µl of 10X TE buffer to the 100µl of eluted DNA. Do not add TE buffer if the DNA is to be used for automated fluorescent sequencing.
- Cap the microcentrifuge tube and store the purified plasmid DNA at –20°C or below.
Ethanol Precipitation
Use Ethachinmate (NIPPON GENE、312-01791).
- Add 3.3 µL of 3M Sodium Acetate (attached with Ethachinmate) into 100µL of DNA solution.
- Add 1µL of Ethachinmate.
- Vortex.
- Add ethanol, 200-250µL.
- Vortex.
- Centrifuge at 12000xg for 5min.
- Precipitation.
Electrophoresis
- Prepare 200mL of a 1.0% agarose solution:
- Measure 2.0g agarose into a beaker.
- Add 200mL 1xTAE buffer.
- Wrap the top of the beaker with plastic wrap.
- Punch a hole through the wrap with a pipette tip (To let out steam).
- Dissolve the agarose by heating in microwave and swirling without boiling.
- Allow the agarose to cool.
- Pour the agarose solution into a gel tray on a gel maker.
- If there is air bubbles, pushing them with a pipette tip.
- Place comb in the maker.
- Cover the maker with a plastic wrap.
- Let stand for about 45min.
- Remove the comb carefully.
- Store in the Tupperware in the refrigerator.
- Place the tray in electrophoresis chamber.
- Cover the tray with 1xTAE buffer.
- To prepare samples for electrophoresis, add 1µL of 6x Loading Buffer for every 5µL of DNA solution and mix well.
- Load 6µL of the DNA solution per well.
- Electrophoresis at 100V for about 30min until Loading Buffer have migrated approximately three-quarters of the gel.
- Stain the gel in 0.5µg/mL ethidium bromide for 20-30min.
- Rinse the gel with MilliQ.
- Place a plastic wrap on the transilluminator in the cabinet of Printgraph.
- Place the gel on the transilluminator.
- Turn on the transilluminator and confirm the position of the gel.
- Shoot the picture.
- Turn off the transilluminator.
- Dispose of the gel.
PCR Purification
Use Wizard SV Gel and PCR Clean-Up System by Promega.
- Combine 1 part sample (PCR product) with one part Membrane Binding Solution (e.g. 50μl sample+ 50μl).
- Apply the solution to the column, and let stand for 1min.
- Centrifuge for 1min at 13000rpm. Discard the flow-through.
- Add 700µl Membrane Wash Solution.
- Centrifuge for 1min and discard the through.
- Add 500μl Membrane Wash Solution.
- Centrifuge for 5min and discard the through.
- Place the column in a clean tube.
- Add 60µl MilliQ to the center of each column, let stand for 1min.
- Centrifuge for 1min at 13000rpm.
- Discard the column.
qRT-PCR
Use QuantiTect SYBR green PCR kit Cat. No. 204143 by QIAGEN
- Dilute primer to 1.5μM.
- Dilute RT products.
- Mix the following;
| 2x QuantiTect SYBR Green PCR Master Mix | 22.5μL |
| 1/20xRT products | 4.5μL |
| MilliQ | 9μL |
| total | 36μL |
- Mix the reaction mix thoroughly, and dispense 36μL into 96 wells plate.
- Add primer set 9μL.
- Mix by inverting and voltex.
- Dispense 10μL into 384 wells plate and centrifuge.
- Let stand for 2min at 50°C and for 15min for 95°C.
- 40 cycles for 15sec at 95°C, for 30sec at 60°C, and for 1min at 72°C.
- Let stand for 15sec at 95°C.
Ligation
- Make 2µL of Mixture (the vector and the insert at 1 : 5-10) and Control (only the vector).
- Add 5µL Ligation High, 1µL T4 Kinase, and 7µL MilliQ to create a solution.
- Incubate at 16℃ for 30 min. If the colonies of E.coli transformed with the Control
Restrictive Digestion
Use EcoRI, XbaI, SpeI, PstI, (NEB)
- Mix the following.
| Sample | 5µL |
| 10xBuffer | 1µL |
| Restriction Enzyme | 0.1µL |
| MilliQ | -3.9µL |
- Let stand for 2h at 37℃
Media
M9 medium
- Stir Na2HPO4 6 g, KH2PO4 3 g, NaCl 0.5 g and NH4Cl 1 g with water 1L.
- After autoclave, add 10 ml filter sterilized 100 mM MgSO4, 20 % glucose, 10 mM CaCl2, and 100 mM thiamine-HCl.
If you need, add 10 ml filter sterilized 20 % casamino acid.
LB medium
- Stir Tryptone 20 g, Yeast extract 10 g and NaCl 10 g with water 200ml.
- If you make LB plates, add agar 10 g.
- Autoclave.
SOB medium
- Stir Tryptone 20g and Yeast extract 5g with water.
- Add 2 ml 5 M NaCl and 840 ul 3 M KCl and add water up to 1L.
- After autoclave, add 10 ml filter sterilized 1 M MgSO4 and 1 M MgCl2.
SOC medium
- Add 2 M glucose 1 ml to 100 ml SOB.
Buffer TB
- Stir PIPES and CaCl2・2H2O with 100 ml water.
- Add 8.315 ml 3 M KCl.
- Add KOH and adjust pH 6.8.
- Add MnCl2・4H2O.
- Add water up to 200 ml.
- Filter sterilize.
Solubilization of Antibiotics
- Mix the following (Final concentration is 50mg/mL).
Ampicillin
| Ampicillin | 1.0g |
| MilliQ | 20mL |
Kanamycin
| Kanamycin | 0.5g |
| MilliQ | 10mL |
- Dispense 1.1mL of the solution into 1.5mL tubes.
- Store in the -20℃ freezer.
Transformation
- Unfreeze conpitent cells on ice.
- Dry a plate by letting the plate upside down and partly open in incubator.
- Add 1µL DNA solution and 20µL competent cells to 1.5mL tube, let stand for 30min on ice. If few colonies are observed, increase the amount of the competent cells or DNA, but make the amount of DNA not to get over that of the competent cells.
- Heatshock for 60s at 42℃.
- Let stand for 2min on ice.
- Culture for 1h in preculture medium (LB or SOC medium), and plate by using spreader. Do not heat spreader too much because E.coli will dead for heat.
Sequence
Use Big Dye Terminator 3.1(ABI)
- Mix the following
| 5xBuffer | 2µL |
| Primer (3.2µM) | 1µL |
| Template Plasmid | 200ng |
| Big Dye Terminator 3.1 | 0.5µL |
| MilliQ | up to 10µL |
- Let stand for 1min at 96℃.
- 35 cycles for 5s at 98℃, for 5s 50℃, and for 2.5min at 68℃.
- Add 25µL 100% ethanol and 1µL NAOAC
Flower Fairy E.coli
Western blotting
Gel solution
| Running gel | Stacking gel | |
|---|---|---|
| 1.5M Tris-HCl(pH8.8) | 2.5mL | - |
| 0.25M Tris-HCl(pH6.8) | - | 3.0mL |
| 30% Acrylamide | 5mL | 0.6mL |
| 10% SDS | 0.2mL | 0.12mL |
| DW | 2.3mL | 2.3mL |
| TEMED | 15µL | 7µL |
| 10% APS | 100µL | 60µL |
| Total | 10mL | 6mL |
4x Sample buffer
| 1M Tris-HCl | 2mL |
| SDS | 0.8g |
| 100% glycerol | 4mL |
| 14.7M mercaptoethanol | 0.4mL |
| 0.5M EDTA | 1mL |
| Bromophenol blue | 8.0mg |
| DW | 2.6mL |
10x electrode buffer
| Tris | 15.15g |
| Glycin | 71.55g |
| 10%SDS | 50mL |
| DW | 450mL |
| Total | 500mL |
blotting buffer
| Tris | 12g |
| Glycin | 14.4g |
| DW | 800mL |
| Methanol | 200mL |
TBST
| 50mM | Tris |
| 150mM | NaCl |
| 0.1% | Tween-20 |
blocking buffer
- TBST
- 5% skim milk
AP color development buffer
- 100mM Tris (pH8.5)
- 100mM NaCl
- 5mM MgCl2
- Spin down 100µL culture and suspend into sample buffer.
- Boil at 95°C for 10 min.
- Apply 10µL to polyacrylamide gel.
- Electrophorese at 500V, 30mA for 50 min.
- Transfer at 50V, 100mA for 30 min.
- Add blocking buffer to the membrane and incubate with shaking for 30 min. at room temperature.
- Add appropriate primary antibody diluted in TBST and incubate with shaking for 30 min. at RT.
- Wash with TBST and incubate with shaking for 10 min, two times.
- Add secondary antibody conjugated with AP and incubate with shaking for 30 min. at RT.
- Wash with TBST and incubate with shaking for 10 min, three times.
- Add 66µL NBT and 33µLof BCIP into color development buffer, and add the mixture to the membrane.
- After the coloring, wash with DW.
Verification of R9 function by GFP
Verification of R9 function by TAKEUCHI
| R9(20µg/µL) | 0.9µL |
| GFP(1.2mg/mL) | 2.23µL |
| RBS | 16.85µL |
| total | 20µL |
X5
Method:
1. Peel cuticles on parafilm by using the head of pencil. (Menasha, wI, 54952)
2. Put plant cells into GFP&R9 or GFP for 5~30min.
3. Put plant cells into PBS.
4. Hoechst dyeing.
| 1 | 2 | 3 | 4 | 5 | 6 | |
| R9 | o | o | o | x | o | o |
| cuticle | o | o | o | o | x | x |
| soak in GFP | 5min | 15min | 30min | 5min | 5min | 30min |
Cultivation of Arabidopsis thaliana
Preparation
- MS culture medium
- mixed salts for MS culture medium
- Vitamin stocked solution(×1000)You can preserve this solution in a refrigerator for a few months.
- Mix the following(以下、表の形式)
| thiamine hydrochloride | 300mg |
| nicotinic acid | 500mg |
| pyridoxine hydrochloride | 50mg |
| DW | up to 100ml |
- sucrose
- 0.1N KOH
- Gellan gum or Gelrite
- sterilized solution (Prepare this solution when you need it.)
- Mix the following
| sodium hypochlorite | 2,5ml |
| Tween 20 | 25μl |
- sterilized water
- sterilized by heating 0.1%(w/v) agar
- 70% ethanol
Condition
- MS culture medium
- light intensity : 3,000 lux
- day length 8 hour light period(10:00 – 18:00) / 16 hour dark period
- temperature 22℃
- 4 seeds / a culture medium
Method
- Put seeds into a 1,5ml microtube.
- Add 1ml of 70% ethanol to it, and stir it well.
- Centrifuge a microtube lightly (6,000 rpm / a few seconds / at room temperature), and dump supernatant.
- Add 1ml of sterilized liquid to it, and stir it with rotater for 3~5 minutes. Work in a clean bench except centrifuging after this manipulation,.
- Centrifuge a microtube lightly, and dump supernatant.
- Add 1ml of sterilized liquid to it, and stir it well.
- Centrifuge a microtube lightly, and dump supernatant.
- Repeat steps of [6. to 7.] 3~5 times.
- Suspend seeds in a few hundreds~ one thousand μl of 0.1% agar solution.
- Take proper quantity of seeds with a micropipette, and drop it on culture medium.
Extruction of protein in E.coli with lysozyme
- Harvest the bacterial cells by centrifugation at 3,000g for 15min at 4℃.
- Resuspend pelleted bacterial cells in the quantity of 1/20 of the culture field of lysis buffer.
- Add 1/50 of the whole of lysozyme solution.
- Mix gently for 30min on ice.
- Add 1/50~1/10 of the whole of 10% Triton X-100.
- Mix for 20 min at 4℃.
- Remove cell debris and unneeded things by centrifugation at 12,000g for 30min at 4℃.
- Collect the clear top of the liquid.
notes
Lysis buffer : 50 mM Tris-HCl or Phosphate, 150 mM NaCl, 1 mM PMSF, pH8.0
Lysozyme solution : 10 mg/ml lysozyme in 20 mM Tris-HCl, pH8.0
Purification of protein with His Tag
- Put 1/10~1/20 of lysate of Ni-NTA resin on column.
- Wash Ni-NTA resin with 3 times of whole amount of column of PBS.
- Equilibrate Ni-NTA resin with 3 times of whole amount of column of Equilibration buffer.
- (Add equilibration buffer to column and cover under nozzle of column with palafilm then leave for 30min in order to drop equilibration buffer)
- Bond proteins with Ni-NTA resin adding lysate to column with equilibrated Ni-NTA resin
- Wash with 10 times of the whole of wash buffer
- Elution with 5 times of the whole of elution buffer
- Collect Fraction by an equal amount
notes
Equilibration buffer : 20mM Tris-HCl(pH7.5) 150mM NaCl
Wash buffer : Equilibration buffer + 20mM imidazole
Elution buffer : Equilibration buffer + 200mM imidazole
Confocal microscope
Making a prepared specimen
- PFA 500μL
- MilliQ 2100μL
- Hoechst 1μL
- Slide glass
- Gather fungus body by centrifugal separation.
- Add 500μL of the PFA to a microtube containing fungus body and resuspend pellet.
- Keep shading it for 10minutes at 4℃.
- Centrifuge it for 1minute at 16,000×g and discard the supernatant.
- Add 1000μL of MilliQ and 1μL of Hoechst to the microtube, resuspend pellet and incubate bacterial suspension for 30minutes or more.
- Centrifuge bacterial suspension for 1minute at 16,000×g and discard the supernatant.
- Add 1000μL of MilliQ, resuspend pellet, centrifuge bacterial suspension and discard the supernatant again.
- Add 100μL of MilliQ and resuspend pellet.
- Pipette bacterial suspension on a slide glass and dry it.
- Preserve it with shading it.
Golden Gate Assembly
Preparation for “Golden Gate Assembly
- Design of primers for PCR
There are some rules in designing primers for PCR, when you make primers to get PCR productions for “Golden Gate Assembly”. Firstly you have to design them as you can add an adapter, which includes BsaI recognition site and BsaI restriction site, to PCR productions. (The BsaI recognition sequence "GGTCTC" is separated from its four bp overhang by a single bp, and BsaI activity is independent of the sequences of the single bp spacer and the four bp overhang.) Secondly you need to design primers as any two parts in a bin are completely interchangeable with respect to Golden Gate assembly, and only a single pair of oligos is required for each part across the entire assembly.
Golden Gate Assembly Protocol
- Add 100 ng of the linearized vector backbone and equimolar amounts of the other assembly pieces to a 15 µl total volume assembly reaction mixture as follows:
| linearized vector backbone | each additional assembly piece | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 Ligase, 2 million cohesive end units / mL | dH2O |
|---|---|---|---|---|---|---|
| 100 ng | to equimolar with backbone | 1.5 µl | 0.15 µl | 1µl | 1 µl | to 15µl |
- NOTE: It is essential to use a High Concentration Ligase
- BsaI is only 10% active at 37 C without the addition of BSA.
- Perform the assembly reaction in a thermocycler as follows:
either (following Engler 2009):
3 min @ 37 C }
4 min @ 16 C } 25 cycles
5 min @ 50 C }
5 min @ 80 C } 1 cycle
- Transform 5 µl of the assembly reaction into 100 µl of competent E. coli and/or run a diagnostic agarose gel to check for successful assembly.
We constructed these BioBrick parts in 2012!!
<groupparts>iGEM012 Kyoto</groupparts>
This year, we define FT protein (BBa_K797006) , FT generator with His-tag (BBa_K797015) , T7-6His-R9 (BBa_ K797014) , RBS-TorA (BBa_K797002) , and Plasmid Backbone for golden gate assembly (BBa_ K797013) as our favorite BioBrick parts of 2012.We confirmed that these parts function properly.
FT protein (BBa_K797006)
FLOWERING LOCUS T(FT) protein, is an identity of florigen, plants' hormone that induce flowering at a low dose. This protein is the transcription factor for promoting the expression of other genes, such as API and FUL.
FT generator with His-tag (BBa_K797015)
This device is composed of T7 promoter (BBa_I712074), RBS and FT gene (BBa_K797006) with Histag. This device doesn't have terminator so that you can combine any part you like, such as GFP, with FT generator.
T7-RBS-6His-R9 (BBa_ K797014)
We can extract proteins by His tag, and penetrate proteins into cells by R9 sequence.
RBS-TorA (BBa_K797002)
This part is coding N-terminal TorA signal region with RBS. Proteins conjugated TorA signal are recognized by Tat secretion pathway and they are secreted from cytoplasm to periplasm while maintaining their holdings. This comes from the part with the enzyme of TMAO reductase. This part includes RBS so that you don't bother to insert new RBS between this signal and your promoter.
Plasmid Backbone for golden gate assembly(BBa_ K797013)
This backbone is suitable for golden gate assembly. Golden gate assembly is a suitable assembly to combine many parts in order you want. This plasmid backbone has restriction enzyme cleavage site of Dpn1, let you build construction by Golden gate assembly in one ligation.
We conducted Golden gate assembly by ourselves and confirmed this part has the restriction cite and restriction enzyme cutting site of Bsa1. Please see detail on Experiment page of this part.
Strains
E.coli
JM109 endA, recA1, gyrA96,thi,hsdR17,(rκ-,mκ+),relA1, supE44,λ-,Δ(lac-proAB),[F',traD36,proAB,laclqZΔM15]
BL21(DE3) F-, ompT, hsdSB(rB -mB - ), gal, dcm, (DE3)
Plant
Arabidopsis thaliana ecotype Columbia
Primers
Florigen
| Name | sequence |
|---|---|
| FT(RBS+)forward | gctctagagaaagaggagaaaGTCGACATGTCTATAAATATAAGAGACCCTCT |
| FT(RBS+)reverse | aaactgcagcggccgctactagtaCCTTGAGCTCCTAAAGTCTTCTTCCTC |
| FT(RBS-)forward | ggaattcgcggccgcttctagagATGTCTATAAATATAAGAGACCCTCTT |
| FT for sequence forward | TGACCATGATTACGCCAAGC |
| FT for sequence reverse | ACGGCCAGTGAATTGTAATACG |
| FT(+His)forward | TCTATAAATATAAGAGACCCTCTTATAGTAAGCAGAGTTG |
| FT(+His)reverse | ATGGTGATGGTGATGGTGCATGTCGACtttctcctctttctctagtatctcc |
| FT mutation forward | CATTCATCGTGTCGTGTTTATATTGTTTCG |
| FT mutation reverse | CCAGCAGTGGGACTTGGATTTTCGTAACAC |
| FT mutation forward2 | CATTCATCGTGTCGTGTTTATATTGTTTCGACAGC |
| TUBULIN forward | AAACTCACTACCCCCAGCTTTG |
| TUBULIN reverse | CACCAGACATAGTAGCAGAAATCAAGT |
| SEP3 forward | CTAAGACTAAGGTTAGCTGATGGGTA |
| SEP3 reverse | ATGATGACGACCGTAGTGATCAA |
| FUL forward | TGCGTAACCTCCTCCAGAGAT |
| FUL reverse | GTTCTACTCGTTCGTAGTGGTAGGAC |
| AP1 forward | ACATCCGCACTAGAAAAAACCAAC |
| AP1 reverse | CTTGGTTCTGCTGATCCCACA |
| pspA forward | ggaattcgcggccgcttctagagCTGGCATGTTGCTGTTGATTC |
| pspA reverse | aactgcagcggccgctactagtaATGCAGCCATAACCAGATCG |
| TatABCD forward | ggaattcgcggccgcttctagagGTCTGATCGCCTGGTTTGTC |
| TatABCD reverse | aactgcagcggccgctactagtaCAGTGTGAAGAATACCGAGTTCC |
| TMAO reductase forward | ggaattcgcggccgcttctagagTAAGGCTCACGGCGATAAGA |
| TMAO reductase reverse | aactgcagcggccgctactagtaCGATTTGCGTCAGTTGTTCA |
| kil forward | ggaattcgcggccgcttctagagTCCAAATGGGATTGCTAGGAC |
| kil reverse | aactgcagcggccgctactagtaGTAGCTCTTGATCCGGCAAAC |
| for sequence | |
| TatABCD forward | tcaggtgtgggcatttatcg |
| TatABCD reverse | gaccacaaagtaggcgaatgc |
| TMAOreductase forward | cgtggatgattgtggttctg |
| TMAOreductase reverse | gtgctgctgtagcctttgtagt |
Golden Gate Assembly
| Name | sequence |
|---|---|
| psB1K3 6’ | tttGGTCTCagttcTACTAGTAGCGGCCGCTGCA |
| psB1K3 1 | tttGGTCTCatggaCTCTAGAAGCGGCCGCGAAT |
| lacI 1’ | tttGGTCTCatccaTGGTGCAAAACCTTTCGC |
| lacI 2 | tttGGTCTCacataGGCGCTATCATGCCATAC |
| GFP 2’ | tttGGTCTCatatgGAGCACTACTAGAGAAAGAG |
| GFP 3 | aaaCTTGTgagaccTGATGCCTGGCTCTAGTATT |
| GFP 5 | tttGGTCTCaccatTGATGCCTGGCTCTAGTATT |
| RFP 3’ | tttGGTCTCacaagCACTACTAGAGAAAGAGGAG |
| RFP 4 | aaaAAGCTgagaccCACTAGCACTATCAGCGTTA |
| RFP 5 | aaaCCATTgagaccCACTAGCACTATCAGCGTTA |
| CFP 4’ | tttGGTCTCagcttATACTGAGCACTACTAGAGA |
| CFP 5 | aaaCCATTgagaccTTGATGCCTGGCTCTAGTAT |
| DT 5’ | tttGGTCTCaatggCCAGGCATCAAATAAAACGA |
| DT 6 | tttGGTCTCagaacTATAAACGCAGAAAGGCCCA |
| J23106 1' | tttGGTCTCagttcTTTACGGCTAGCTCAGTCCT |
| J23106 2 | tttGGTCTCatggaGCTAGCACTATACCTAGGACT |
| J23106 2' | tttGGTCTCatccaTTTACGGCTAGCTCAGTCCT |
| J23106 3 | tttGGTCTCacataGCTAGCACTATACCTAGGACT |
| J23106 3' | tttGGTCTCatatgTTTACGGCTAGCTCAGTCCT |
| J23106 4 | tttGGTCTCacttgGCTAGCACTATACCTAGGACT |
| J23106 4' | tttGGTCTCacaagTTTACGGCTAGCTCAGTCCT |
| J23106 5 | tttGGTCTCaaagcGCTAGCACTATACCTAGGACT |
| araC 5' | tttGGTCTCagcttGCGTAACAAAAGTGTCTATAATCACGG |
| araC 6 | tttGGTCTCaccatATGGAGAAACAGTAGAGAGTTGCGATA |
| GFP(sequence) rev | CACCCTCTCCACTGACAG |
| GFP(sequence) Fw | CAGCTGCTGGGATTACAC |
| RFP(sequence) rev | CATGAACTCTTTGATAACGTCTTC |
| RFP(sequence) Fw | CAGTACGAACGTGCTGAAG |
| DT(sequence) rev | GAAGGTGAGCCAGTGTGA |
| DT(sequence) Fw | TCTGTTGTTTGTCGGTGAAC |
 "
"