Team:Potsdam Bioware/Project/Potsdam Standard
From 2012.igem.org
(→Experimental design) |
(→Potsdam Standard - BBF_RFC_91: Phosphorothioate-based BioBrick cloning Standard) |
||
| (43 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class="box_round gradient_grey"> | <div class="box_round gradient_grey"> | ||
| - | + | ==Potsdam Standard - BBF_RFC_91: Phosphorothioate-based BioBrick cloning Standard== | |
| - | + | ||
| - | + | [[media:UP12_BBF RFC 91 phosphorothioate based cloning.pdf|download > 'BBF RFC 91 phosphorothioate based cloning.pdf']] | |
| - | === | + | |
| + | ===Problem description=== | ||
<br> | <br> | ||
While using the common assembly standards to assemble different parts we noticed that these procedures are very time and material consuming and therefore difficult for long and different assemblies. Another problem is the usage of restriction enzymes and gel separation in every step of the cloning process because of different conditions and efficiencies for different enzymes and the danger of mutation after UV-exposure in the gel. | While using the common assembly standards to assemble different parts we noticed that these procedures are very time and material consuming and therefore difficult for long and different assemblies. Another problem is the usage of restriction enzymes and gel separation in every step of the cloning process because of different conditions and efficiencies for different enzymes and the danger of mutation after UV-exposure in the gel. | ||
| + | The greatest problem are undesired restriction sites in your gene of interest. Therefore, you have to mutate these site before using your desired restriction enzymes. | ||
<br><br> | <br><br> | ||
| - | === | + | |
| + | ===Solution: the Potsdam Assembly Standard=== | ||
<br> | <br> | ||
| - | The Potsdam Assembly Standard is a modified PLICing (Phosphorothioate-based ligase-independent gene cloning) method (Blanusa ''et al.'' (2010)). For | + | The Potsdam Assembly Standard is a modified PLICing (Phosphorothioate-based ligase-independent gene cloning) method (Blanusa ''et al.'' (2010), Liu XP ''et al.'' (2010)). The original PLICing is based on the amplification of insert and backbone to ligate them together by knocking out thiophosphates at the 3' end (Figure 1). For the Potsdam Standard, we designed a new cloning vector with an RFP expression cassette as insert and two new restriction enzyme recognition sites in the suffix and prefix in pSB1C3. In the prefix we added the Apa I recognition site and in the suffix the Sph I recognition site. Both enzymes causing a 3’ overhang with 4 nucleotides. For the cloning process we cut the vector with Apa I and Sph I. In this assembly standard that’s the only step where we have to use restriction enzymes.<br> |
The insert was amplified with primers that contain 4 phosphothioate nucleotides and the recognition sites for Apa I and Sph I at the 5’ end. After incubation in an iodine/ethanol solution the thiophosphates were cut out resulting in a 3’ overhang which is suitable to the overhangs that was created by cutting the new assembly vector. | The insert was amplified with primers that contain 4 phosphothioate nucleotides and the recognition sites for Apa I and Sph I at the 5’ end. After incubation in an iodine/ethanol solution the thiophosphates were cut out resulting in a 3’ overhang which is suitable to the overhangs that was created by cutting the new assembly vector. | ||
After that the digested vector and the PLICed insert were mixed and transformed into <i>E. coli</i>. By using the RFP expression cassette we created a ligation control system. Due to the fact that red fluorescent colonies have a failed vector ligation we can tell which colonies are correctly ligated. The colonies that do not show red fluorescence are the positive clones.<br><br> | After that the digested vector and the PLICed insert were mixed and transformed into <i>E. coli</i>. By using the RFP expression cassette we created a ligation control system. Due to the fact that red fluorescent colonies have a failed vector ligation we can tell which colonies are correctly ligated. The colonies that do not show red fluorescence are the positive clones.<br><br> | ||
| + | [[file:UP12_PLICing_theory.jpg|400px|center|thumb|'''Figure 1:''' PLICing method published by Blanusa ''et al.'' 2010]] | ||
| - | === | + | ===Experimental design=== |
<br> | <br> | ||
| - | Firstly, we amplified our gene of interest (GOI) with primers which had an thiophosphate overhang. | + | Firstly, we amplified our gene of interest (GOI) with primers which had an thiophosphate overhang. The overhang for the forward primer has to be GGCCT to inactive the Apa I restriction site and for the reverse primer CATGA to inactivate the Sph I restriction site after ligation. These overhangs are complementary to the digested restriction sites of Apa I and Sph I in the new cloning RFP standard vector. For the PCR we used the Phusion polymerase. After the PCR, we pliced the GOI with 100 mM iodine solution in 99 % ethanol and the 0.5 M Tris-HCl cleavage buffer with pH = 9.0. For doing this, we mixed 8 µL GOI with 1 µL cleavage buffer, 0.4 µL Milli-Q water and 0.6 µL iodine solution. After that we incubated the reaction mixture 5 min at 70 °C using a thermocycler. After the plicing process, the pliced GOI was mixed with the digested backbone and ligated at room temperature for 1 h and directly transformed into ''E. coli'' (summary: figure 2). |
<br><br> | <br><br> | ||
| - | [[file: | + | [[file:PotsdamStandardscheme.jpg|600px|center|thumb|'''Figure 2:''' Experimental design of the Potsdam Standard]] |
| - | === | + | ===Results=== |
<br> | <br> | ||
| - | + | ||
| - | + | ====For culturing ''E.coli'' cells transformed with Potsdam Standard Cloning Vector with RFP==== | |
<br> | <br> | ||
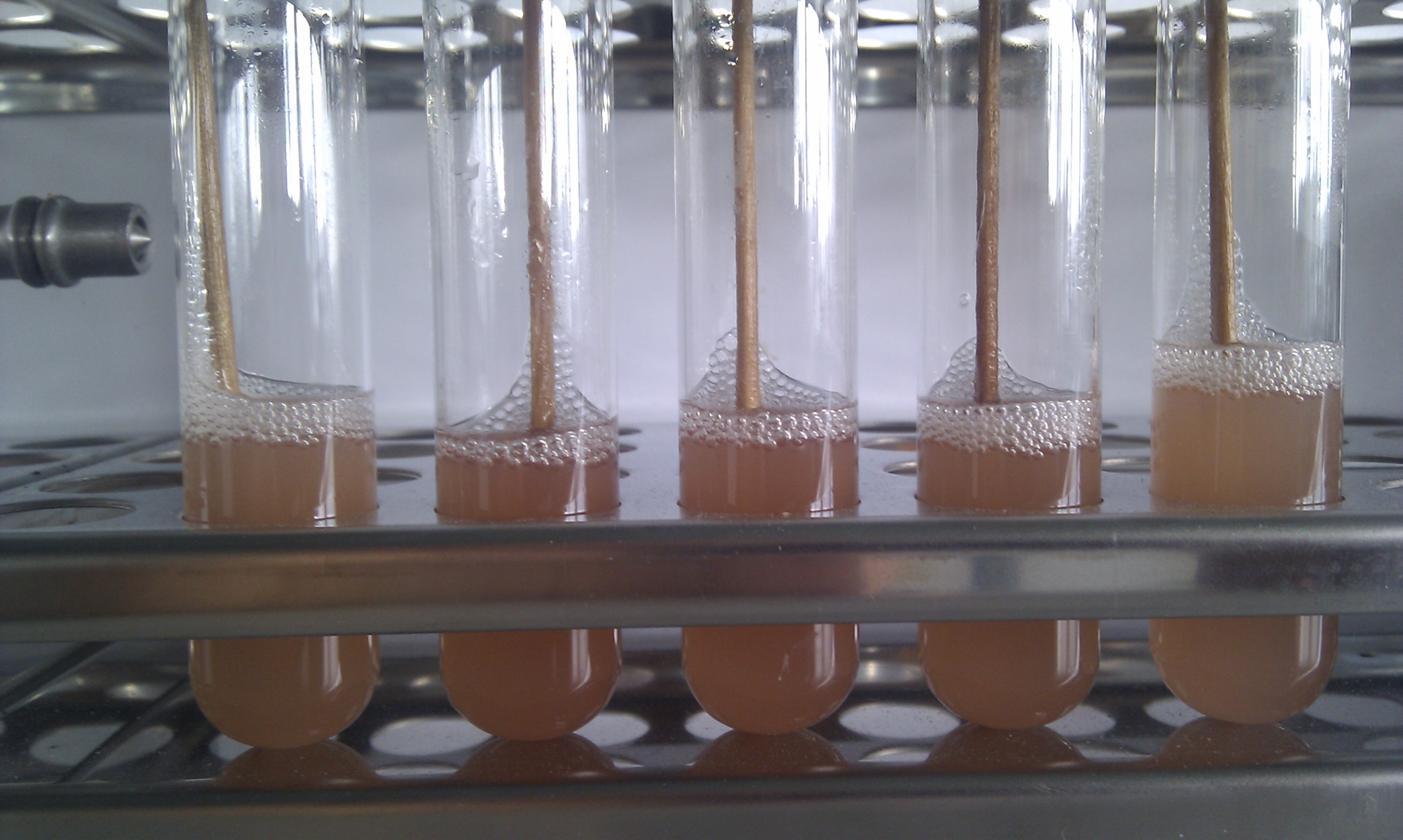
| - | + | The red fluorescence of the transformed colonies is obvious after 24 h incubation at 37 °C and over night incubation at 8 °C. To speed up the cloning, you can use LED light to see red fluorescence even after 16 h incubation at 37 °C. The transformed ''E.coli'' cells do not need any IPTG for RFP production. For the overnight culture (figure 3), we observed that it is better to incubate them with IPTG. After centrifugation, you can see the cherry-coloured cell pellets (figure 4).<br> | |
<br> | <br> | ||
| - | + | ||
| + | ====Using Potsdam Standard Cloning Vector with RFP as ligation control==== | ||
<br> | <br> | ||
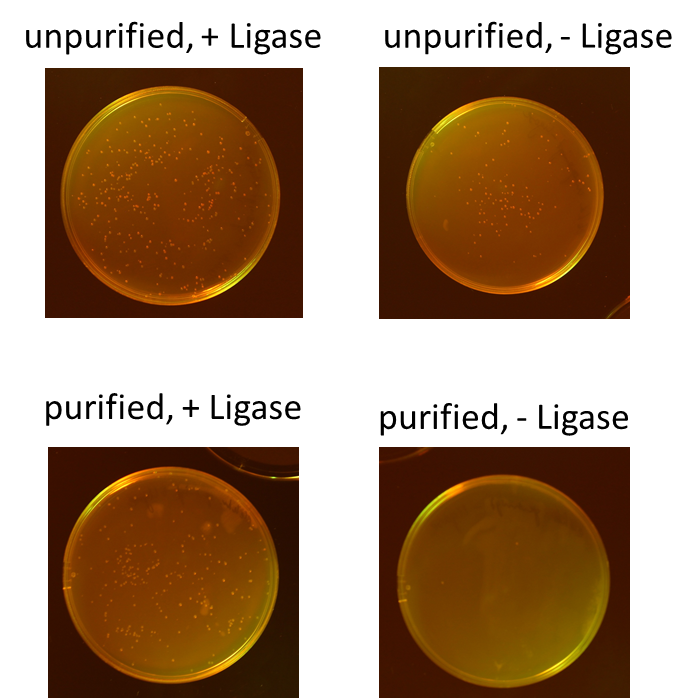
| - | + | We tested different combination to prove the most efficient experimental design. Therefore, we tried four different conditions: the ligation of the pliced GOI with the unpurified digested backbone with and without ligase and the ligation of the pliced GOI with the purified digested backbone with and without ligase. After transformation, we picked only clones which have lost their red fluorescence indicating that the ligation was successful (figure 5).<br> | |
| + | To quantify the results, we counted the red colonies and the colonies without red fluorescence (figure 6). It is obvious, that the ligation with the electrophoretic purified digested backbone is more efficient than without any purification. Nevertheless, the use of ligase is not necessary for the ligation process under these conditions.<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <table class="table center" border=0><tr><td>[[file:RFP_culture.jpg|400px|center|thumb|'''Figure 3:''' Overnight culture of transformed ''E.coli'']]</td><td>[[File:UP12_RFP_pellets.jpg|400px|center|thumb|'''Figure 4:''' After centrifugation, the cell pellets are coloured like cherries]]</td><tr><td>[[file:UP12_Ligation.png|400px|center|thumb|'''Figure 5:''' Results of different ligation conditions]]</td><td>[[file:UP_12_Potstdam_Standard.png|400px|center|thumb|'''Figure 6:''' Results of different ligation conditions]]</td></tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ===References=== | ||
| + | <br> | ||
| + | * Blanusa M, Schenk A, Sadeghi H, Marienhagen J, Schwaneberg U., Phosphorothioate-based ligase-independent gene cloning (PLICing): An enzyme-free and sequence-independent cloning method. Anal Biochem, 2010 Nov 15;406(2):141-6. [http://www.ncbi.nlm.nih.gov/pubmed/20646988|''Pubmed PMID: 20646988''] | ||
| + | * Liu XP, Liu JH, The terminal 5' phosphate and proximate phosphorothioate promote ligation-independent cloning. Protein Sci. 2010 May;19(5):967-73. [http://www.ncbi.nlm.nih.gov/pubmed?term=the%20terminal%205'%20phosphate%20and%20proximate%20phosphorothioate%20promote|''Pubmed PMID: 20217896''] | ||
| + | |||
</div> | </div> | ||
Latest revision as of 02:14, 27 October 2012
Contents |
Potsdam Standard - BBF_RFC_91: Phosphorothioate-based BioBrick cloning Standard
download > 'BBF RFC 91 phosphorothioate based cloning.pdf'
Problem description
While using the common assembly standards to assemble different parts we noticed that these procedures are very time and material consuming and therefore difficult for long and different assemblies. Another problem is the usage of restriction enzymes and gel separation in every step of the cloning process because of different conditions and efficiencies for different enzymes and the danger of mutation after UV-exposure in the gel.
The greatest problem are undesired restriction sites in your gene of interest. Therefore, you have to mutate these site before using your desired restriction enzymes.
Solution: the Potsdam Assembly Standard
The Potsdam Assembly Standard is a modified PLICing (Phosphorothioate-based ligase-independent gene cloning) method (Blanusa et al. (2010), Liu XP et al. (2010)). The original PLICing is based on the amplification of insert and backbone to ligate them together by knocking out thiophosphates at the 3' end (Figure 1). For the Potsdam Standard, we designed a new cloning vector with an RFP expression cassette as insert and two new restriction enzyme recognition sites in the suffix and prefix in pSB1C3. In the prefix we added the Apa I recognition site and in the suffix the Sph I recognition site. Both enzymes causing a 3’ overhang with 4 nucleotides. For the cloning process we cut the vector with Apa I and Sph I. In this assembly standard that’s the only step where we have to use restriction enzymes.
The insert was amplified with primers that contain 4 phosphothioate nucleotides and the recognition sites for Apa I and Sph I at the 5’ end. After incubation in an iodine/ethanol solution the thiophosphates were cut out resulting in a 3’ overhang which is suitable to the overhangs that was created by cutting the new assembly vector.
After that the digested vector and the PLICed insert were mixed and transformed into E. coli. By using the RFP expression cassette we created a ligation control system. Due to the fact that red fluorescent colonies have a failed vector ligation we can tell which colonies are correctly ligated. The colonies that do not show red fluorescence are the positive clones.
Experimental design
Firstly, we amplified our gene of interest (GOI) with primers which had an thiophosphate overhang. The overhang for the forward primer has to be GGCCT to inactive the Apa I restriction site and for the reverse primer CATGA to inactivate the Sph I restriction site after ligation. These overhangs are complementary to the digested restriction sites of Apa I and Sph I in the new cloning RFP standard vector. For the PCR we used the Phusion polymerase. After the PCR, we pliced the GOI with 100 mM iodine solution in 99 % ethanol and the 0.5 M Tris-HCl cleavage buffer with pH = 9.0. For doing this, we mixed 8 µL GOI with 1 µL cleavage buffer, 0.4 µL Milli-Q water and 0.6 µL iodine solution. After that we incubated the reaction mixture 5 min at 70 °C using a thermocycler. After the plicing process, the pliced GOI was mixed with the digested backbone and ligated at room temperature for 1 h and directly transformed into E. coli (summary: figure 2).
Results
For culturing E.coli cells transformed with Potsdam Standard Cloning Vector with RFP
The red fluorescence of the transformed colonies is obvious after 24 h incubation at 37 °C and over night incubation at 8 °C. To speed up the cloning, you can use LED light to see red fluorescence even after 16 h incubation at 37 °C. The transformed E.coli cells do not need any IPTG for RFP production. For the overnight culture (figure 3), we observed that it is better to incubate them with IPTG. After centrifugation, you can see the cherry-coloured cell pellets (figure 4).
Using Potsdam Standard Cloning Vector with RFP as ligation control
We tested different combination to prove the most efficient experimental design. Therefore, we tried four different conditions: the ligation of the pliced GOI with the unpurified digested backbone with and without ligase and the ligation of the pliced GOI with the purified digested backbone with and without ligase. After transformation, we picked only clones which have lost their red fluorescence indicating that the ligation was successful (figure 5).
To quantify the results, we counted the red colonies and the colonies without red fluorescence (figure 6). It is obvious, that the ligation with the electrophoretic purified digested backbone is more efficient than without any purification. Nevertheless, the use of ligase is not necessary for the ligation process under these conditions.
References
- Blanusa M, Schenk A, Sadeghi H, Marienhagen J, Schwaneberg U., Phosphorothioate-based ligase-independent gene cloning (PLICing): An enzyme-free and sequence-independent cloning method. Anal Biochem, 2010 Nov 15;406(2):141-6. [http://www.ncbi.nlm.nih.gov/pubmed/20646988|Pubmed PMID: 20646988]
- Liu XP, Liu JH, The terminal 5' phosphate and proximate phosphorothioate promote ligation-independent cloning. Protein Sci. 2010 May;19(5):967-73. [http://www.ncbi.nlm.nih.gov/pubmed?term=the%20terminal%205'%20phosphate%20and%20proximate%20phosphorothioate%20promote|Pubmed PMID: 20217896]
 "
"