Team:Colombia/Project/Experiments/Our Design
From 2012.igem.org
(→Our design) |
|||
| (20 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{https://2012.igem.org/User:Tabima}} | {{https://2012.igem.org/User:Tabima}} | ||
| + | <div class="right_box"> | ||
| + | =Our design= | ||
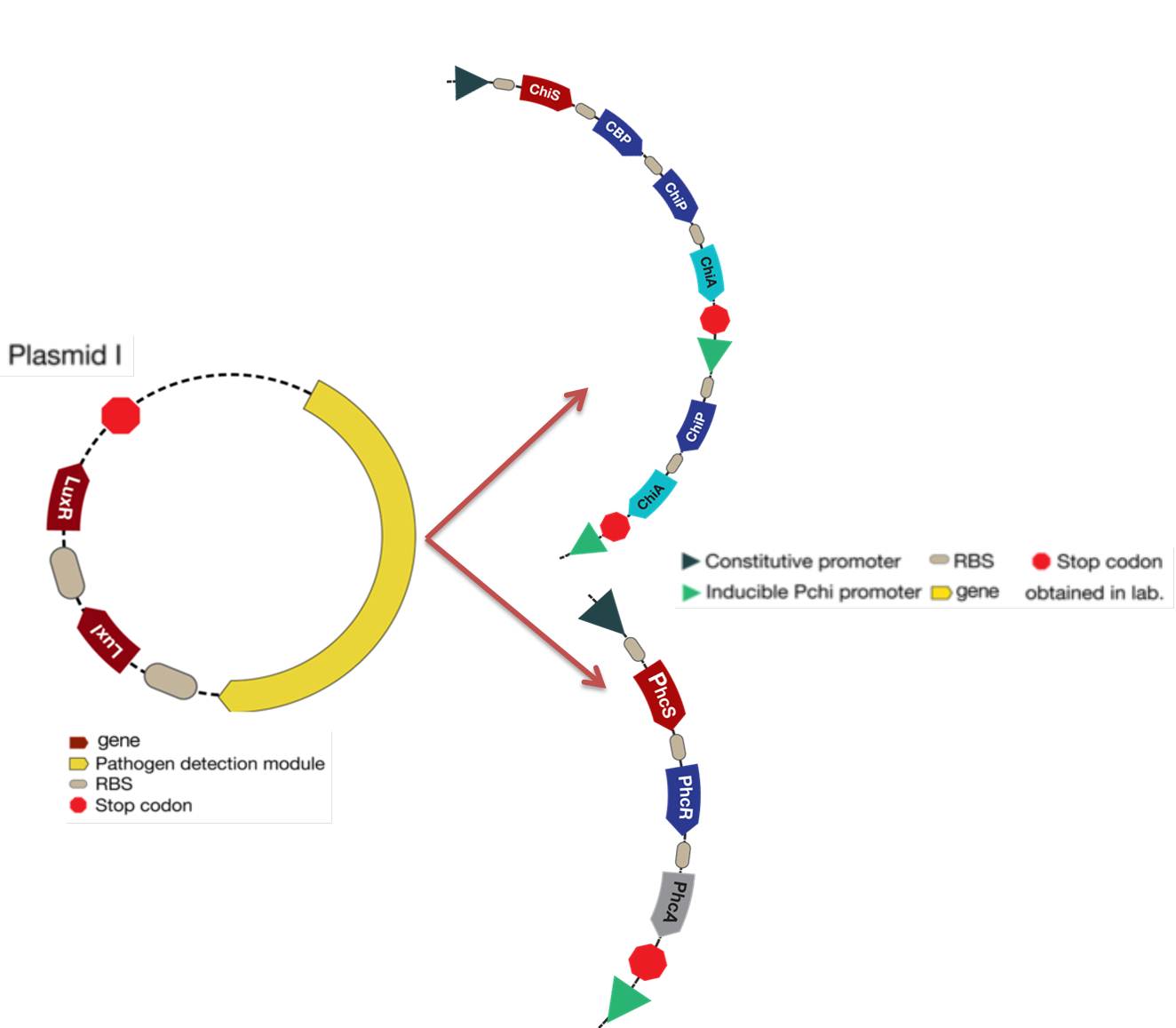
| - | [[File: | + | The purpose of our design is to generate genetically-modified bacteria with a "detect and alert" system. With this aim, we designed two plasmids: |
| + | <html> | ||
| + | <br> | ||
| + | </br> | ||
| + | </html> | ||
| + | -The first plasmid has two parts, one part is changeable and the other one is always present. The changeable part is the pathogen detection module that changes depending on the organism that is going to be detected. This year we have worked in the detection of [https://2012.igem.org/Team:Colombia/Project/Experiments/Aliivibrio_and_Streptomyces fungus (chitin detection)] or [https://2012.igem.org/Team:Colombia/Project/Experiments/Ralstonia ''Ralstonia solanacearum''(3 OH-PAME detection)], so detection module has that two options. Furthermore, the detection turns on the second plasmid (Plux promoter) by generating a signal (LuxI-LuxR complex), doesn’t matter which pathogen we want to detect the signal generated is always same because this part doesn’t change. | ||
| + | |||
| + | |||
| + | [[File:modelocorr.jpg|650px|thumb|center|Figure 1. Detection module located in the plasmid I.]] | ||
| + | |||
| + | |||
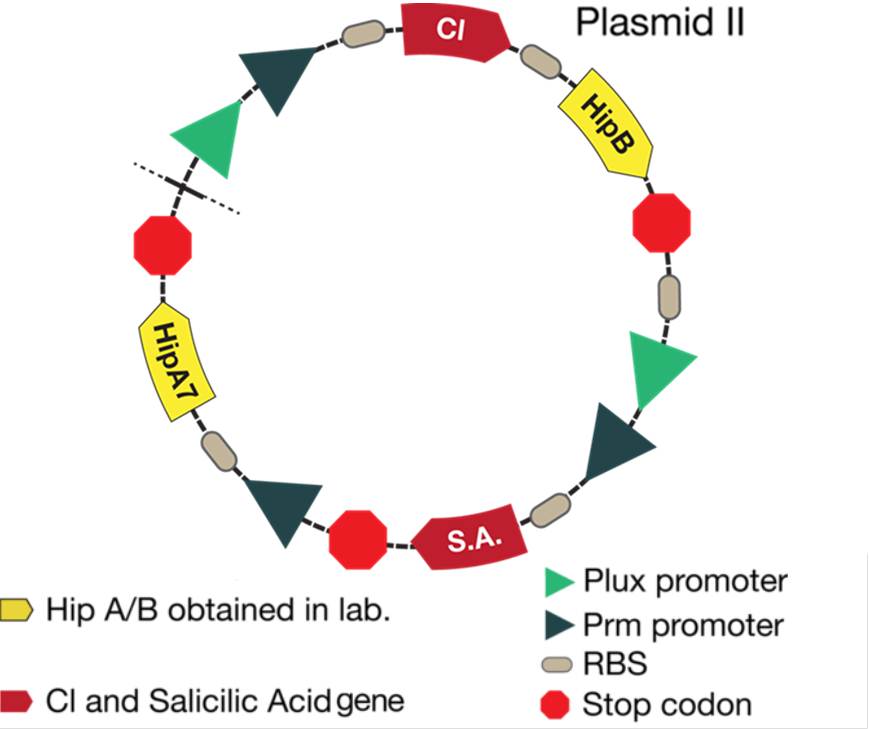
| + | -The second plasmid never changes because it´s aim is to alert the plant and ¨sleep¨ the bacteria. For alert the plant, is produced salicylic acid that activates the plant's defense trough a systemic acquired reponse and for control the bacterial density, we introduced a [https://2012.igem.org/Team:Colombia/Project/Experiments/Pseudomonas toxin-antitoxin system (HipA-HipB)] that allows to reduce the quantity of active population when the pathogen is not detected and to control the bacterial population growth. | ||
| + | |||
| + | |||
| + | [[File:p2corr.png|500px|thumb|center|Figure 2. Alert system and toxin/antitoxin system located in the plasmid II.]] | ||
| + | |||
| + | |||
| + | Our design was checked through [https://2012.igem.org/Team:Colombia/Modeling/Diff mathematical models], that helped us to make sure that our design works and even it helped us to change some parts from the first design proposed. With this mathematical models we can predict what is going to happen when our designed bacteria is working with the plants. | ||
| + | |||
| + | </div> | ||
Latest revision as of 02:50, 27 October 2012
Template:Https://2012.igem.org/User:Tabima
Our design
The purpose of our design is to generate genetically-modified bacteria with a "detect and alert" system. With this aim, we designed two plasmids:
-The first plasmid has two parts, one part is changeable and the other one is always present. The changeable part is the pathogen detection module that changes depending on the organism that is going to be detected. This year we have worked in the detection of fungus (chitin detection) or Ralstonia solanacearum(3 OH-PAME detection), so detection module has that two options. Furthermore, the detection turns on the second plasmid (Plux promoter) by generating a signal (LuxI-LuxR complex), doesn’t matter which pathogen we want to detect the signal generated is always same because this part doesn’t change.
-The second plasmid never changes because it´s aim is to alert the plant and ¨sleep¨ the bacteria. For alert the plant, is produced salicylic acid that activates the plant's defense trough a systemic acquired reponse and for control the bacterial density, we introduced a toxin-antitoxin system (HipA-HipB) that allows to reduce the quantity of active population when the pathogen is not detected and to control the bacterial population growth.
Our design was checked through mathematical models, that helped us to make sure that our design works and even it helped us to change some parts from the first design proposed. With this mathematical models we can predict what is going to happen when our designed bacteria is working with the plants.
 "
"