Team:SJTU-BioX-Shanghai/Project
From 2012.igem.org
(→Overall project) |
Huanan1991 (Talk | contribs) (→Reference) |
||
| (106 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Template:12SJTU_header}} | |
| + | {{Template:12SJTU_nav_project}} | ||
| + | <table><tr> | ||
| + | <td valign="top" width="200"> | ||
| + | {{Template:12SJTU_floatnav_head}} | ||
<html> | <html> | ||
| - | < | + | <script type="text/javascript"> |
| - | < | + | // <![CDATA[ |
| - | + | var myMenu; | |
| - | + | window.onload = function() { | |
| - | + | myMenu = new SDMenu("my_menu"); | |
| - | + | myMenu.oneSmOnly = true; | |
| - | + | myMenu.remember = false; | |
| - | + | myMenu.init(); | |
| - | + | myMenu.expandMenu(myMenu.submenus[0]); | |
| - | + | ||
| - | </ | + | }; |
| + | // ]]> | ||
| + | </script> | ||
</html> | </html> | ||
| + | {{Template:12SJTU_floatnav_project}} | ||
| - | + | {{Template:12SJTU_floatnav_foot}} | |
| + | <td valign="top" width="750"> | ||
| + | __NOTOC__ | ||
| + | <!----------------------------------------------------从这里开始写wiki---------------------------------> | ||
| + | =Project Overview= | ||
| + | {{Template:12SJTU_part_summary_head}} | ||
| - | + | Advance in molecular cloning technology has made it possible for mankind to entitle engineered organisms to different biochemical reactions. However, the speed of those enzymatic reactions is often limited because intermediates produced from upstream enzyme cannot be passed efficiently to downstream enzyme due to spatial obstacles. Thus, synthetic scaffold built to decrease distance between enzymes for speeding biochemical reactions is a rising topic with promising application prospect. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Moreover, although some progress has been made in fields of metabolic flux control with synthetic scaffold, these strategies remain non-dynamic. Artificially and dynamically controlling metabolic flux has remained a challenge. | ||
| + | {{Template:12SJTU_part_summary_foot}} | ||
| - | == | + | ==Introduction== |
| - | + | In this year, we expanded the definition of ''scaffold'' in synthetic biology and developed two universal devices called ''Membrane Accelerator'' and ''Membrane Rudder'' respectively. Together, they made ''Membrane Magic'' happen! | |
| - | + | ||
| - | + | Previous researchers have focused on building protein, RNA or DNA scaffold as constitutive assemblies carrying enzymes. They have succeeded in increasing product yields. However, the amount of those scaffolds could be limited by its expression or copy level, leading to restriction on further acceleration. With ''Membrane Magic'', we made ''E.coli'' membrane into a huge scaffold accommodating enzymes without limitation of scaffold amount. Moreover, protein assembly on membrane could readily receive extracellular or intracellular signal, so the whole system becomes highly tunable. The superiority of Membrane Scaffold is shown in details in '''WHY MEMBRANE'' section below. | |
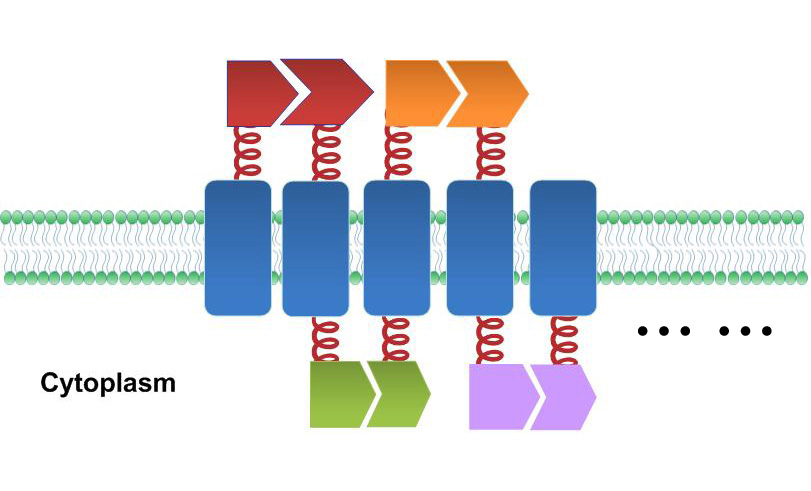
| + | One of our devices, called ''Membrane Accelerator'', functions by localizing and organizing enzymes on membrane surface. ''E.coli'' inner membrane serves as a two-dimensional plane that can accommodate various protein assemblies linked with enzymes. Otherwise diffusing enzymes can form clusters on membrane through interacting protein domains and ligands. Enzyme clusters help substrates flow between enzymes, and thus increase yields of sequential biological reactions. We not only applied the ''Membrane Accelerator'' into biosynthetic pathway but also biodegradation pathway, which is proposed for the first time in synthetic biology. Previous researches on synthetic scaffold controlling metabolic flux all focused on biosynthesis. | ||
| + | [[File:12SJTU membrane accelerator sketch.jpg|thumb|400px|center|''Fig.1:'' Sketch of ''Membrane Accelerator'']] | ||
| + | Although some work has been done in reaction acceleration, it has always been a challenge to artificially and dynamically control those reactions. Our ''Membrane Rudder'' device, however, offers a novel method to control the direction of biochemical reactions through varieties of signals. We further combined the whole post-translational control system with genetic circuits by recruiting RNA aptamer and its corresponding binding protein. Thus RNA signal could also be recruited to dynamically control biochemical reaction. | ||
| + | [[File:12SJTU_overview4.gif|center|400px]] | ||
| - | + | <center>''Fig.2:'' Sketch of ''Membrane Rudder''</center> | |
| - | + | <br\> | |
| - | + | <br\> | |
| - | + | <br\> | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Why MEMBRANE?== | ||
| + | Why do we choose membrane as our primary scaffold to assemble enzymes? | ||
| + | {| | ||
| + | |- | ||
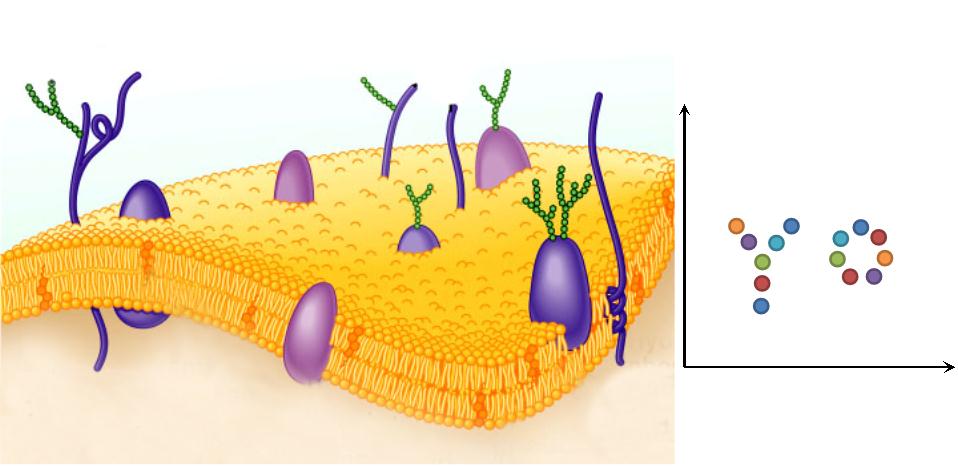
| + | |1. '''Natural Scaffold: ''' Different from previous synthesized scaffold, membrane scaffold is an innate one. Besides, there is no limitation on scaffold amount. | ||
| + | |[[File:12SJTU Why membrane1.png|300px|right|thumb|''Fig.3:'' Natural Scaffold]] | ||
| + | |- | ||
| + | |2. '''Two-Dimensional Plane: ''' Membrane Scaffold changes restricted the reaction space to a two-dimensional plane compared to discrete scaffold. Thus proteins on membrane are more likely to interact with each other (Demonstrated in [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project2.2 Fatty Acid Synthesis:The Refinement of Interaction]). Moreover, we can organize enzymes in 2D pattern on membrane to further facilitate metabolic flux (Demonstrated in [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project2.3 DBT desulfurization]). | ||
| + | |[[File:12SJTU Why membrane2.jpg|300px|right|thumb|''Fig.4:'' Two-Dimensional Plane]] | ||
| + | |- | ||
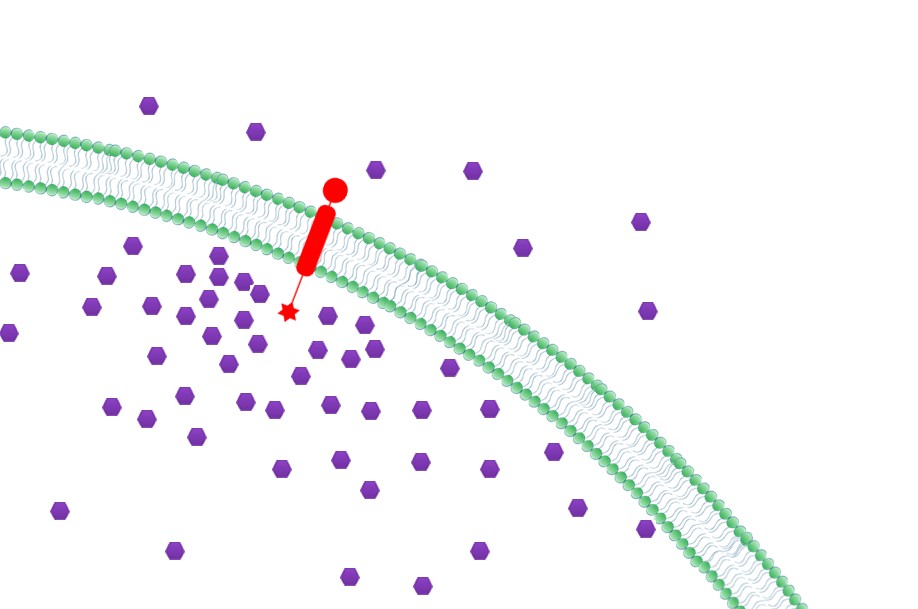
| + | |3. '''Priority to Exportation: ''' Concentration of final products could be effectively increased near the membrane with Membrane Scaffold, which in turn, facilitates the transmembrane transportation. Thus final products would be more readily to be exported to extracellular media. (Demonstrated in [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project2.2 Fatty Acid Synthesis:The Priority to Exportation]) | ||
| + | |[[File:12SJTU Why membrane3.jpg|300px|right|thumb|''Fig.5:'' Priority to Exportation]] | ||
| + | |- | ||
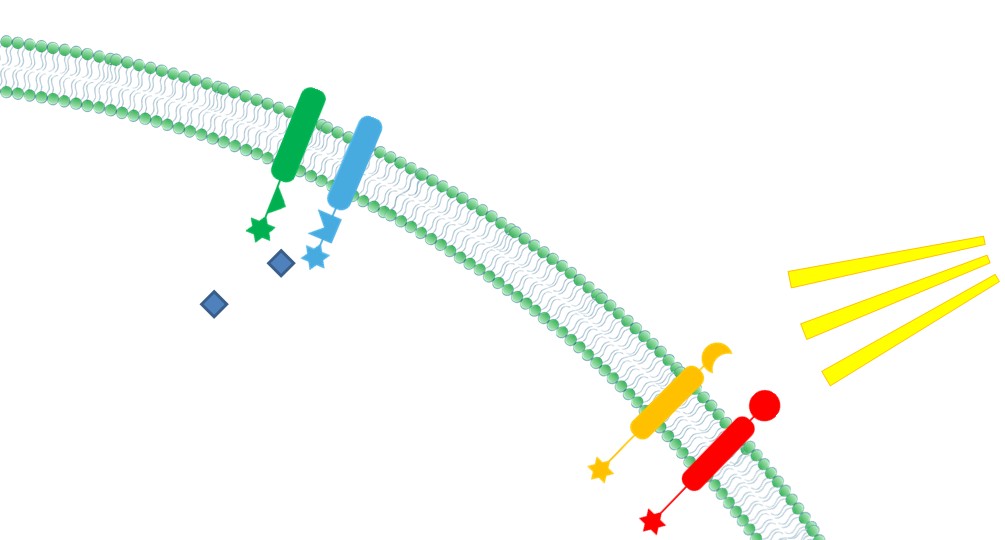
| + | |4. '''Ability to Sense Signals: ''' Membrane Scaffold provides a platform to directly receive environmental and internal signal, So biochemical reactions could be dynamically controlled through those signals(Demonstrated in [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project1.3 Membrane Rudder Design] and [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project2.1 Violacein pathway: Membrane Rudder Application]). | ||
| + | |[[File:12SJTU Why membrane4.jpg|300px|right|thumb|''Fig.6:'' Ability to sense signals]] | ||
| + | |- | ||
| + | |} | ||
| + | ==Reference== | ||
| + | 1.Delebecque, C. J., A. B. Lindner, et al. (2011). "Organization of intracellular reactions with rationally designed RNA assemblies." Science 333(6041): 470. | ||
| - | + | 2.Dueber, J. E., G. C. Wu, et al. (2009). "Synthetic protein scaffolds provide modular control over metabolic flux." Nature biotechnology 27(8): 753-759. | |
| + | 3.Conrado, R. J., G. C. Wu, et al. (2012). "DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency." Nucleic acids research 40(4): 1879-1889. | ||
| + | <!----------------------------------------------------到这里结束---------------------------------------> | ||
| + | </td> | ||
| + | </tr></table> | ||
| - | + | {{Template:12SJTU_footer}} | |
Latest revision as of 04:00, 27 October 2012
| ||||||||||
|
 "
"