Team:Colombia/Project/Experiments/Ralstonia
From 2012.igem.org
m (→Response of PxpsR to synthetic 3 OH-PAME) |
m (→Construction of the PxpsR Reporter) |
||
| (82 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{https://2012.igem.org/User:Tabima}} | {{https://2012.igem.org/User:Tabima}} | ||
| - | = | + | <div class="right_box"> |
| - | + | ||
| - | + | ||
| - | ==PxpsR | + | =''Ralstonia'' Experiments= |
| - | In order to quantify the responsiveness of PxpsR to the presence of 3 OH-PAME we generated a PxpsR-eYFP reporter. The reporter was tested in the pSB1A2 however the part was ensambled into de pSB1C3 backbone and | + | ==''Ralstonia'' BioBricks== |
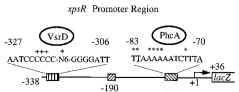
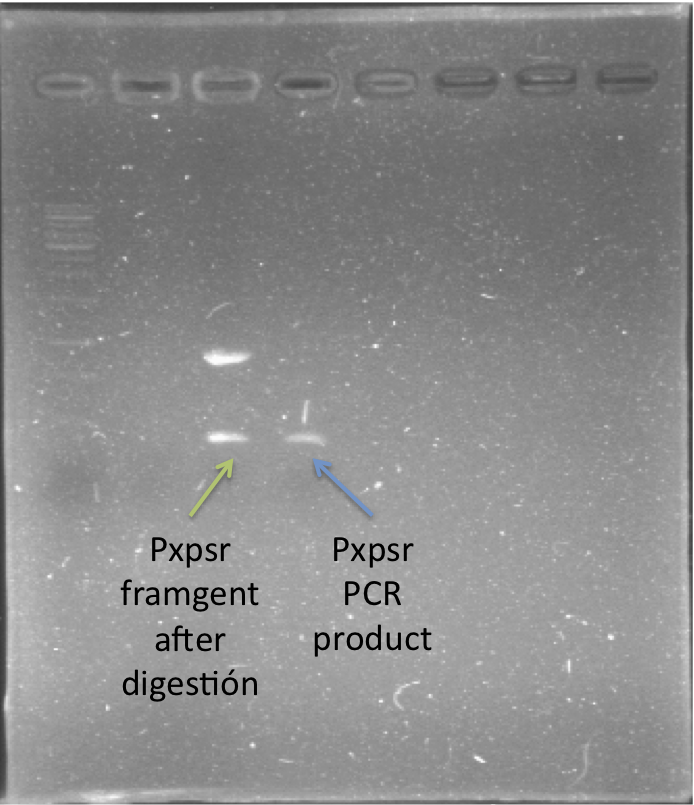
| + | We cloned and generated a biobrick([http://partsregistry.org/Part:BBa_K831016 K831016]) for the promoter region of xpsR (PxpsR). The product of this gene is an integrator signal that regulates the expression of several virulence factors in ''Ralstonia solanacearum'' (Shell, 1996). One of the signals that XpsR integrates is phcA which is involved in the Quorum Sensing mediated by 3 OH-PAME exclusive of ''R. solanacearum''. We designed primers to amplify the upstream region of xpsR gene including the two binding boxes of phcA, the -35 and -10 conserved boxes and excluding the native RBS (Figure 1). This part was confirmed by sequencing. In addition, we cloned the three genes (phcA, phcS and phcR) involved in the sensing of the 3 OH-PAME (Genin and Denny, 2012), further confirmation of these parts is still needed. Image 1 shows the enzymatic confirmation for PxpsR in the backbone pBS1C3. For a more extensive and more detailed description of the cloning procedures, please visit: [https://2012.igem.org/Team:Colombia/Notebook/Journal ''Ralstonia Journal''] | ||
| + | |||
| + | [[File:Captura_de_pantalla_2012-09-26_a_las_16.55.14.png|300px|thumb|center|''Figure 1.''''' xpsR Promoter Region''' Modified from Huang ''et al.'', 1998. Figure shows the boxes upstream in the promoter region of xpsR gene.]] | ||
| + | |||
| + | [[File:ConfpxpsrdigxbaIspeI.png|250px|thumb|center|''Image 1''. '''Confirmation of PxpsR BioBrick'''. Lane 1, MW (Fermentas 1KB ladder). Lane 2, PxpsR cloned in pBS1C3 digest with XbaI and SpeI. Lane 3, Positive control of PxpsR (PCR product).''Note'': We used a reclycled gel to optimize the laboratory reagents.]] | ||
| + | |||
| + | ==Construction of the PxpsR Reporter== | ||
| + | In order to quantify the responsiveness of PxpsR to the presence of 3 OH-PAME we generated a PxpsR-eYFP reporter [http://partsregistry.org/Part:BBa_K831017 (K831017)] using the E0430 BioBrick (Image 2). For a more extensive and more detailed explanation of the cloning procedures, please visit: [https://2012.igem.org/Team:Colombia/Notebook/Journal ''Ralstonia Journal'']. | ||
| + | |||
| + | [[File:PxpsR-eYFP.png|250px|thumb|center|''Image 2''. '''Confirmation of PxpsR-eYFP construct'''. After confirmation of colonies by PCR, plasmids of 2 candidates were digest with EcoRI and SpeI. Lane 1, MW (Fermentas 1KB ladder). Lane 2, Candidate 1: promotorless eYFP (E0430). Lane 3, Candidate 2: PxpsR-eYFP.''Note'': We used a reclycled gel to optimize the laboratory reagents.]] | ||
| + | |||
| + | The reporter was tested in the pSB1A2 however the part was ensambled into de pSB1C3 backbone and sent to the iGEM Registry Parts. Here we show the basal activity (in absence of 3 OH-PAME) of the PxpsR-eYFP reporter (Image 3) and the [http://partsregistry.org/Part:BBa_K831017 functionality]. | ||
| + | |||
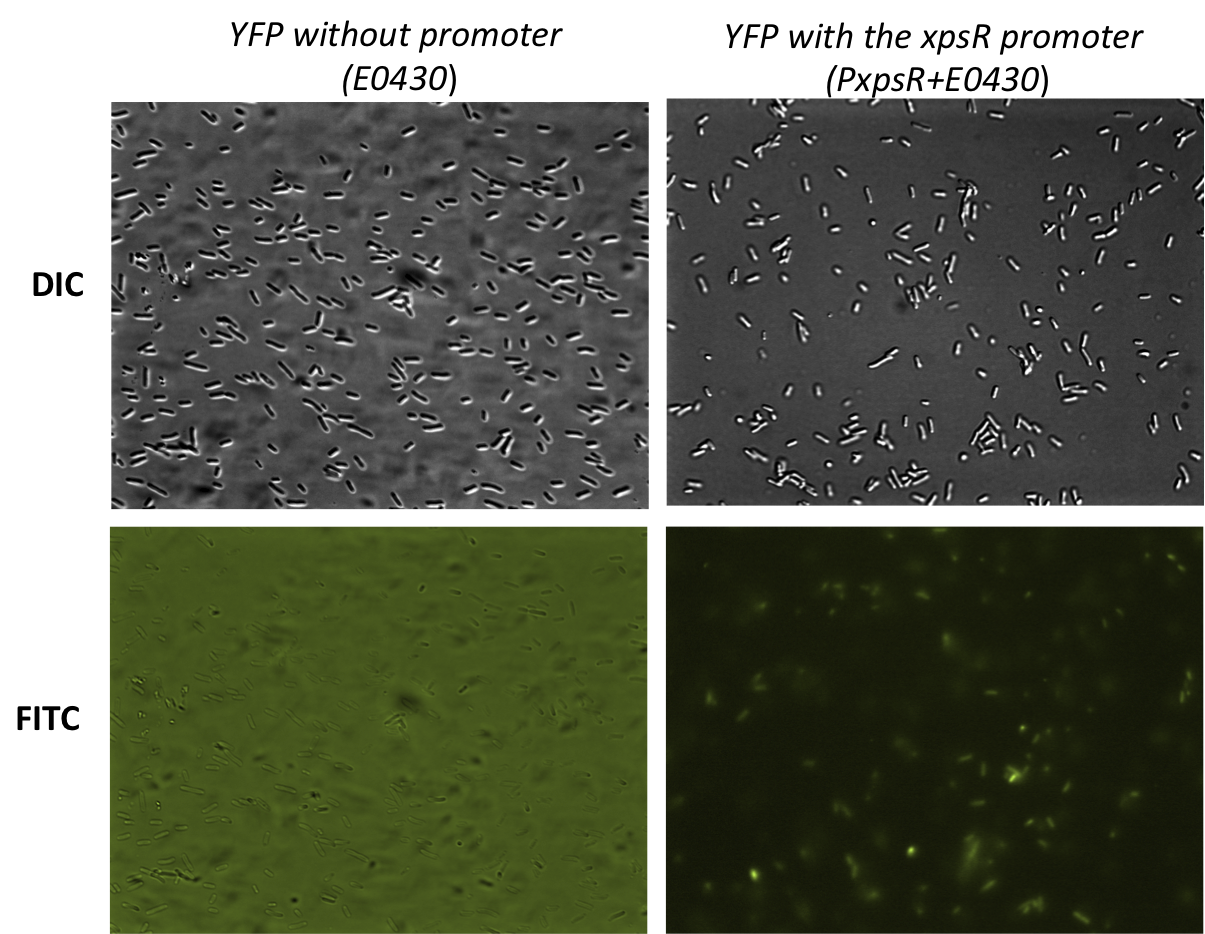
| + | [[File:PxpsR-YFP.png|450px|thumb|center|''Image 3''. '''Functionality of the reporter PxpsR-eYFP'''. Bacterials strains were photographed while using Differential Interference Contrast (DIC) microscopy and Fluorescence microscopy using FITC filter. Bacteria transformed with PxpsR-eYFP construct show basal levels of expression of the reporter while none of the bacteria transformed with the promotorless eYFP show fluorescence. High background levels in the promotorless eYFP are due to the autoflorescence of the LB medium and the high exposition.]] | ||
==Response of PxpsR to synthetic 3 OH-PAME== | ==Response of PxpsR to synthetic 3 OH-PAME== | ||
'''Objective''' | '''Objective''' | ||
| - | To determine the response of PxpsR in a mute ''Ralstonia | + | To determine the induced response of PxpsR in a mute ''Ralstonia solanacearum'' with the presence of synthetic 3 OH-PAME at different concentrations, using eYFP as a fluorescent reporter. |
'''Description''' | '''Description''' | ||
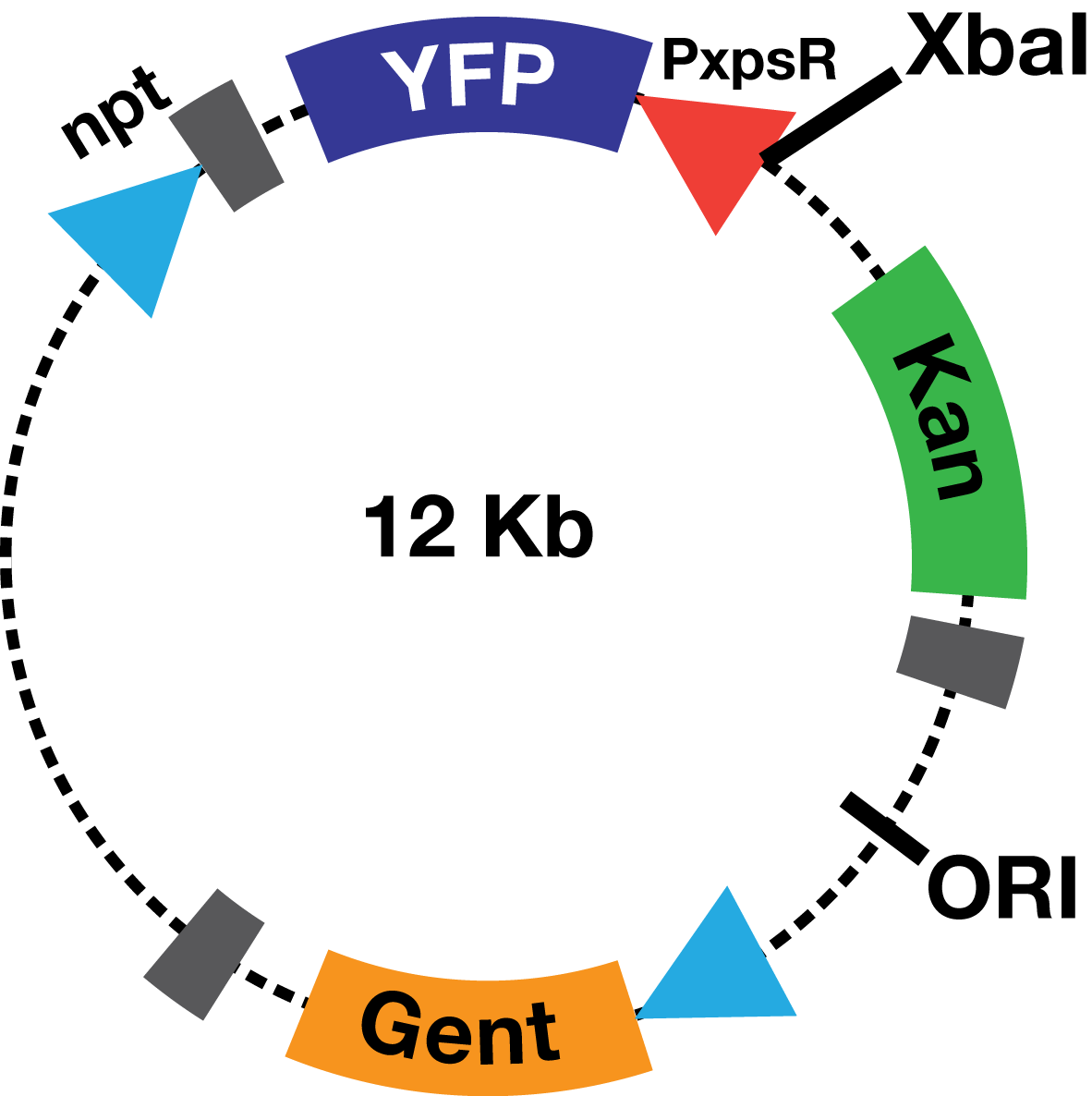
| - | The PxpsR-eYFP biobrick was digested with XbaI and SpeI, and then was ligated to an XbaI digested | + | The PxpsR-eYFP biobrick was digested with XbaI and SpeI, and then was ligated to an XbaI digested ML123 plasmid. This one has a replication origin derived from pVS1, which allows replication in ''Xanthomonas'', ''Pseudomonas'' and ''Ralstonia''. To determine whether the biobrick had ligated in the opposite direction to the strong promoter npt (Figure 2), transformed colonies were confirmed via enzymatic digestions. This way the biobrick would not be induced by the npt promoter present in the plasmid, and therefore, the fluorescent response due to the presence of 3OH-PAME would depend solely on PxpsR. |
| + | |||
| + | [[File:PlasmidoPaolasm.png|200px|thumb|center| '' Figure 2''. '''Scheme of the PxpsR-eYFP reporter in pML123'''.]] | ||
| + | |||
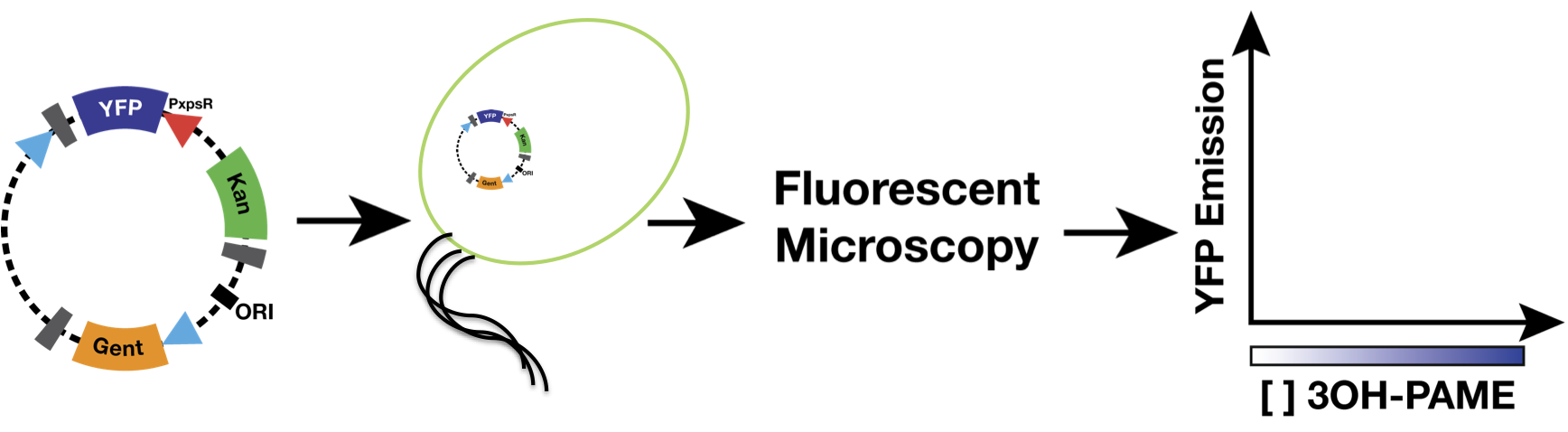
| + | We used as host a mute mutant of ''Ralstonia Solanacearum'', which has a mutation in the gene phcB (3 OH-PAME synthetase). Although it has the ability to sense 3 OH-PAME, it cannot produce it (Figure 3), which allows to create an environment with a known concentration of 3 OH-PAME at all times, indepent of the number of bacterias present. We are currently transforming the mute strain with the PxpsR-eYFP reporter in pML123, once we obtain the transformants we will determine the fluorescent response at different concentrations of synthetic 3-OH-PAME (Figure 4). Based on the fluorescent response, we will be able to determine the inducibility of PxpsR. | ||
| + | |||
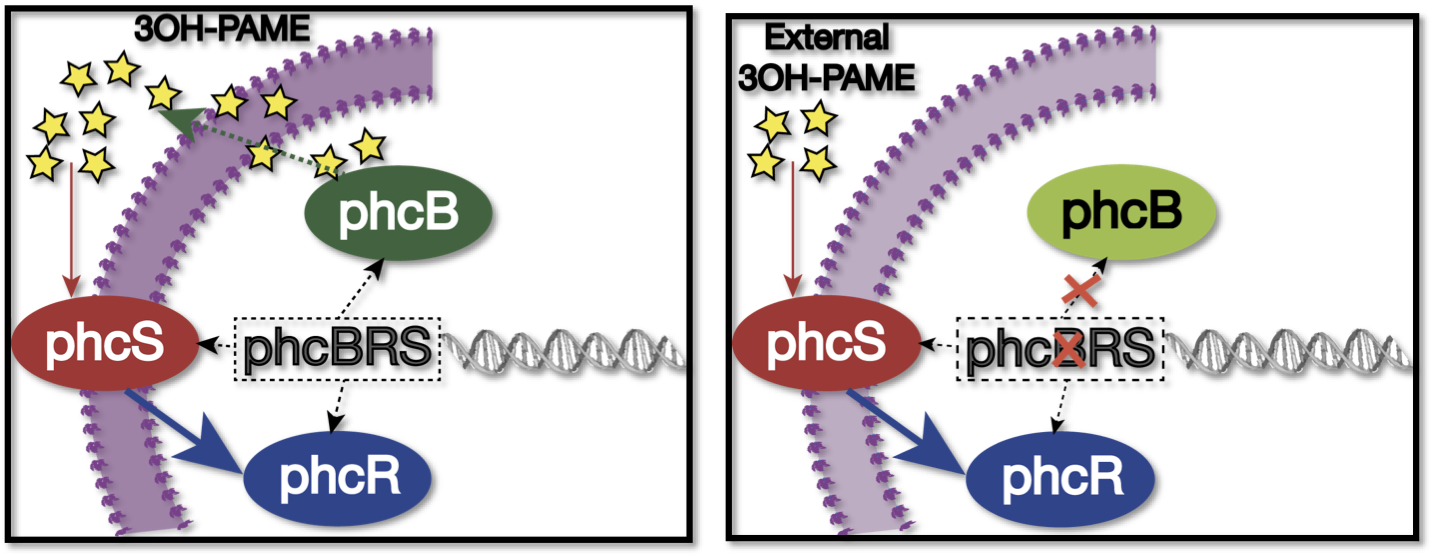
| + | [[File:RalstoniaFigure2.png|500px|thumb|center|''Figure 3''. '''Sensing of 3 OH-PAME'''. Left: When normal sensing of the QS signal, the operon phcBRS regulates the expresion of phcS(sensor), phcR(regulator, quencher of the transcription factor phcA) and phcB (3 OH-PAME synthetase), signal produced diffuses to the environment, while phcS senses the exterior signal. Right: When phcB is mutated, bacterium can sense the external input, but can comunicated with other cells as long as it can't produce 3 OH-PAME.]] | ||
| + | |||
| + | [[File:MethodsRalstonia.png|700px|thumb|center|''Figure 4''. '''Experiment carried out to determine the cuantitative induction of the promoter PxpsR'''.]] | ||
| + | |||
| + | =References= | ||
| - | + | #Schell, M. A. (1996) To be or not to be: how'' Pseudomonas solanacearum'' decides whether or not to express virulence genes. Eur. J. Plant Pathol. 102:459–469. | |
| + | #Genin S. and Denny T.P. (2012) Pathogenomics of the ''Ralstonia solanacearum'' Species Complex. Ann. Rev. Phytopath., 50: 67 -89 | ||
| + | #Huang J., Yindeeyoungyeon W., Garg R.P., Denny T.P. and Schell M.A. (1998) Joint Transcriptional Control of xpsR, the Unusual Signal Integrator of the ''Ralstonia solanacearum'' Virulence Gene Regulatory Network, by a Response Regulator and a LysR-Type Transcriptional Activator. J. Bacteriol. 180(10): 2736-2743 | ||
| - | + | </div> | |
Latest revision as of 01:24, 27 September 2012
Template:Https://2012.igem.org/User:Tabima
Contents |
Ralstonia Experiments
Ralstonia BioBricks
We cloned and generated a biobrick([http://partsregistry.org/Part:BBa_K831016 K831016]) for the promoter region of xpsR (PxpsR). The product of this gene is an integrator signal that regulates the expression of several virulence factors in Ralstonia solanacearum (Shell, 1996). One of the signals that XpsR integrates is phcA which is involved in the Quorum Sensing mediated by 3 OH-PAME exclusive of R. solanacearum. We designed primers to amplify the upstream region of xpsR gene including the two binding boxes of phcA, the -35 and -10 conserved boxes and excluding the native RBS (Figure 1). This part was confirmed by sequencing. In addition, we cloned the three genes (phcA, phcS and phcR) involved in the sensing of the 3 OH-PAME (Genin and Denny, 2012), further confirmation of these parts is still needed. Image 1 shows the enzymatic confirmation for PxpsR in the backbone pBS1C3. For a more extensive and more detailed description of the cloning procedures, please visit: Ralstonia Journal
Construction of the PxpsR Reporter
In order to quantify the responsiveness of PxpsR to the presence of 3 OH-PAME we generated a PxpsR-eYFP reporter [http://partsregistry.org/Part:BBa_K831017 (K831017)] using the E0430 BioBrick (Image 2). For a more extensive and more detailed explanation of the cloning procedures, please visit: Ralstonia Journal.
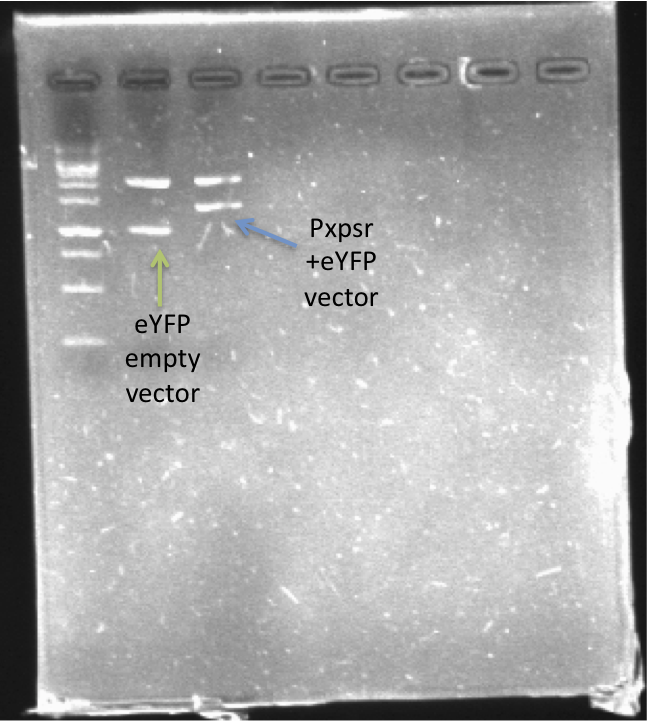
The reporter was tested in the pSB1A2 however the part was ensambled into de pSB1C3 backbone and sent to the iGEM Registry Parts. Here we show the basal activity (in absence of 3 OH-PAME) of the PxpsR-eYFP reporter (Image 3) and the [http://partsregistry.org/Part:BBa_K831017 functionality].
Response of PxpsR to synthetic 3 OH-PAME
Objective To determine the induced response of PxpsR in a mute Ralstonia solanacearum with the presence of synthetic 3 OH-PAME at different concentrations, using eYFP as a fluorescent reporter.
Description The PxpsR-eYFP biobrick was digested with XbaI and SpeI, and then was ligated to an XbaI digested ML123 plasmid. This one has a replication origin derived from pVS1, which allows replication in Xanthomonas, Pseudomonas and Ralstonia. To determine whether the biobrick had ligated in the opposite direction to the strong promoter npt (Figure 2), transformed colonies were confirmed via enzymatic digestions. This way the biobrick would not be induced by the npt promoter present in the plasmid, and therefore, the fluorescent response due to the presence of 3OH-PAME would depend solely on PxpsR.
We used as host a mute mutant of Ralstonia Solanacearum, which has a mutation in the gene phcB (3 OH-PAME synthetase). Although it has the ability to sense 3 OH-PAME, it cannot produce it (Figure 3), which allows to create an environment with a known concentration of 3 OH-PAME at all times, indepent of the number of bacterias present. We are currently transforming the mute strain with the PxpsR-eYFP reporter in pML123, once we obtain the transformants we will determine the fluorescent response at different concentrations of synthetic 3-OH-PAME (Figure 4). Based on the fluorescent response, we will be able to determine the inducibility of PxpsR.
References
- Schell, M. A. (1996) To be or not to be: how Pseudomonas solanacearum decides whether or not to express virulence genes. Eur. J. Plant Pathol. 102:459–469.
- Genin S. and Denny T.P. (2012) Pathogenomics of the Ralstonia solanacearum Species Complex. Ann. Rev. Phytopath., 50: 67 -89
- Huang J., Yindeeyoungyeon W., Garg R.P., Denny T.P. and Schell M.A. (1998) Joint Transcriptional Control of xpsR, the Unusual Signal Integrator of the Ralstonia solanacearum Virulence Gene Regulatory Network, by a Response Regulator and a LysR-Type Transcriptional Activator. J. Bacteriol. 180(10): 2736-2743
 "
"