Team:Bielefeld-Germany/Results/CBD
From 2012.igem.org
(→Cloning of the Cellulose Binding Domains) |
(→Cellulose Binding Domain - Introduction) |
||
| (9 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
| - | In the field of cheap protein-extraction cellulose binding domains (CBD) have made | + | In the field of cheap protein-extraction cellulose binding domains (CBD) have made themselves a name. A lot of publications have been made, concerning a cheap strategy to capture a protein from the cell-lysate with a CBD-tag. Also enhanced segregation with CBD-tagged proteins have been observed. Here the idea is different, we want to take advantage of the binding capacity of binding domains not only for purification reasons (it is still a benefit), but also as an immobilizing-protocol for our laccases. |
To make a purification and immobilization-tag out of a protein domain, there are a lot of decisions and characterizations you have to get through. | To make a purification and immobilization-tag out of a protein domain, there are a lot of decisions and characterizations you have to get through. | ||
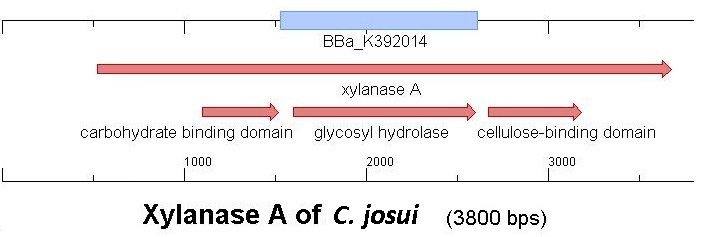
| - | [[Image:Bielefeld2012 Osaka3.jpg| | + | [[Image:Bielefeld2012 Osaka3.jpg|450px|thumb|right|Figure 1:Graphical sequence-alignment of <partinfo>K392014</partinfo> and the [http://www.ncbi.nlm.nih.gov/nuccore/AB041993 Xyn10A gene of C.josui]]] |
| - | Starting with the choice of the binding domain, the first limitation is accessibility. Our first place to look was, of course the [http://partsregistry.org | + | Starting with the choice of the binding domain, the first limitation is accessibility. Our first place to look was, of course the [http://partsregistry.org Partsregistry]. We found a promising Cellulose binding motif of the ''C. josui'' Xyn10A gene (<partinfo>BBa_K392014</partinfo>) there and ordered it right from the spot for our project. After some research later concerning the sequence of that BioBrick it turned out that the part is not the CBD of the Xylanase as it should be, but the glycosyl hydrolase domain of the protein (Figure 1). This result made the part useless for our project ([http://partsregistry.org/Part:BBa_K392014:Experience complete review]) and it was the only binding domain in the [http://partsregistry.org Partsregistry] that fitted to our project. |
| - | So we started to search the accessible organisms we had via NCBI for binding-domains, -proteins and -motifs and asked in work-groups if they could help us out. The results of our database research were only two chitin/carbohydate binding | + | So we started to search the accessible organisms we had via NCBI for binding-domains, -proteins and -motifs and asked in work-groups if they could help us out. The results of our database research were only two chitin/carbohydate binding modules within the ''Bacillus halodurans'' genome (we ordered that stain for it's laccase <partinfo>BBa_K863020</partinfo>). One is in the [http://www.ncbi.nlm.nih.gov/nucleotide/289656506?report=genbank&log$=nuclalign&blast_rank=1&RID=0JPT9WMS01N Cochin chitinase gene] and the other in a [http://www.ncbi.nlm.nih.gov/protein/BAB05022.1 chitin binding protein]. |
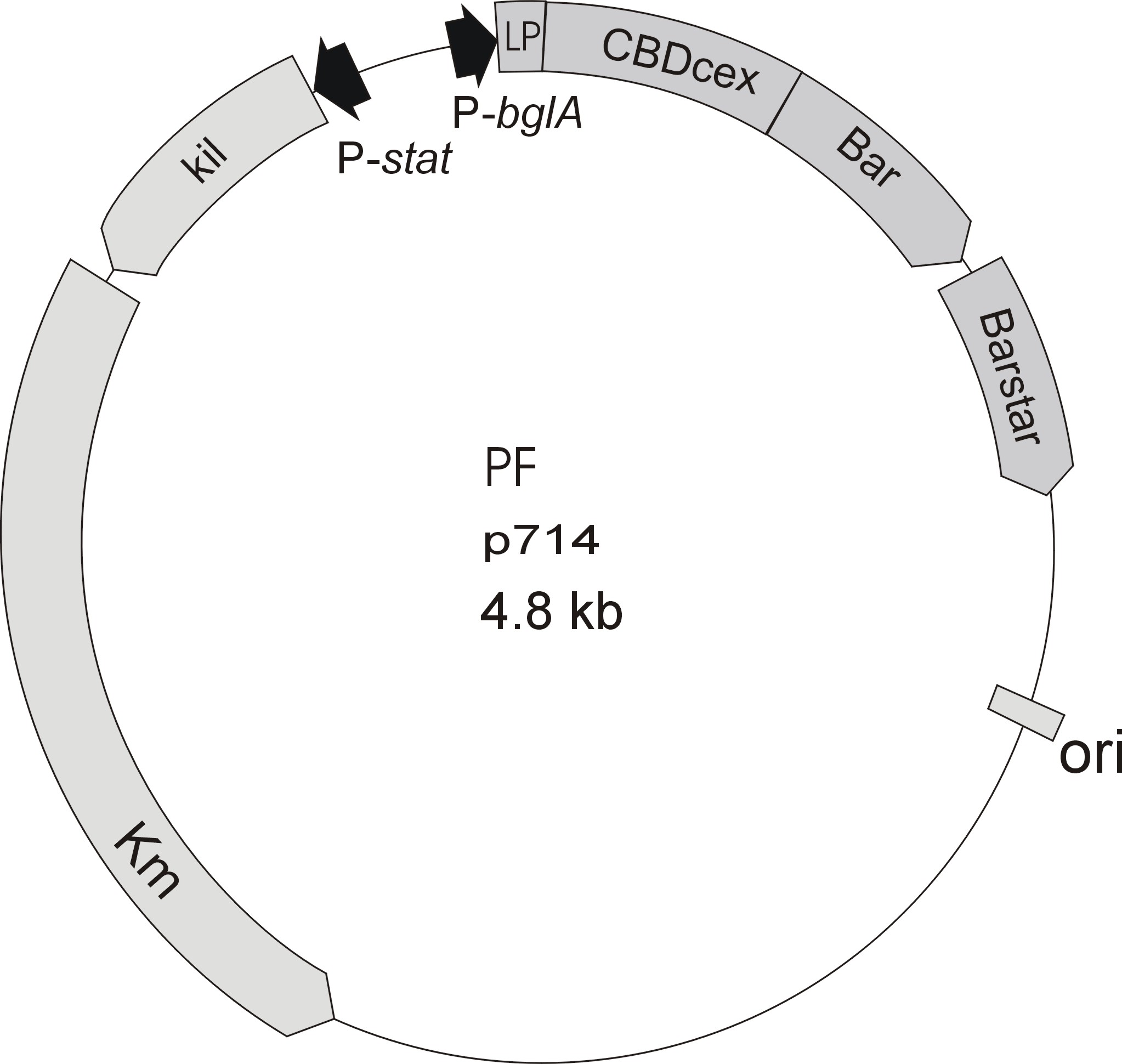
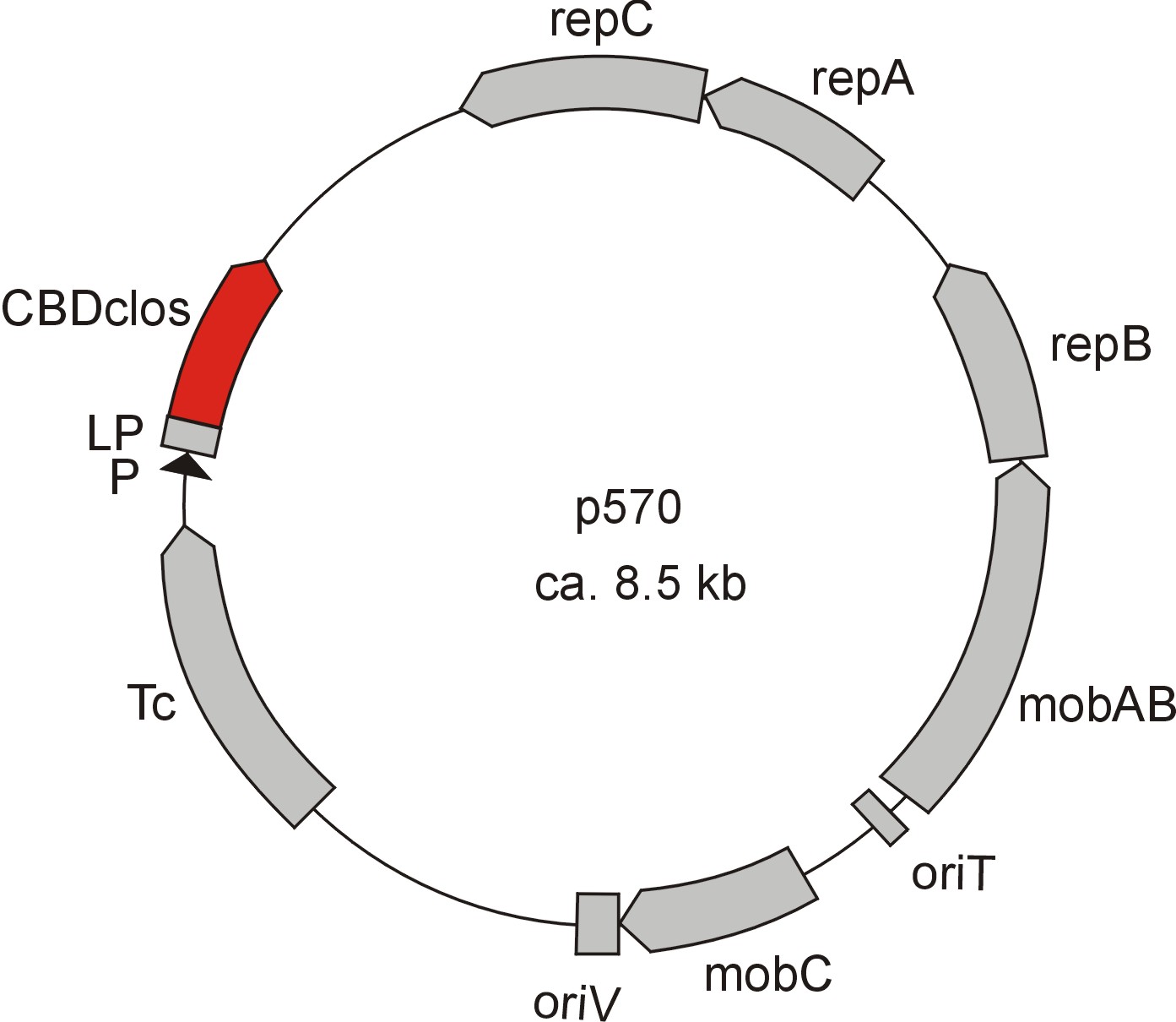
| - | [[Image:Bielefeld2012_p714.jpg| | + | [[Image:Bielefeld2012_p714.jpg|210px|thumb|left|Figure 2: Plasmid with CBDcex]][[Image:Bielefeld2012_p570.jpg|230px|thumb|left|Figure 3: Plasmid with CBDclos]] |
| - | Meanwhile the Fermentation group offered use two plasmids (p570 & p671), containing two different cellulose binding domains. The Cellulose binding domain of the [http://www.ncbi.nlm.nih.gov/nucleotide/327179207?report=genbank&log$=nucltop&blast_rank=3&RID=152ZCN0E01N ''Cellulomonas fimi'' ATCC 484 exoglucanase gene] (CBDcex) and the Cellulose binding domain of [http://www.ncbi.nlm.nih.gov/nuccore/M73817 ''Clostridium cellulovorans'' cellulose binding protein gene (cbp A)] (CBDclos). At that point we decided to use these two domains. Staying within the cellulose binding domain-family and leave other protein domains like carbohydrate binding domains aside will keep the results comparable. Like changing to a different binding material would change the binding capacities of both domains in the same way. Also we would stay within bacterial CBD and didn't have to spend time thinking about post-translational modification and | + | Meanwhile the Fermentation group of our university offered use two plasmids (p570 & p671), containing two different cellulose binding domains. The Cellulose binding domain of the [http://www.ncbi.nlm.nih.gov/nucleotide/327179207?report=genbank&log$=nucltop&blast_rank=3&RID=152ZCN0E01N ''Cellulomonas fimi'' ATCC 484 exoglucanase gene] (CBDcex) and the Cellulose binding domain of [http://www.ncbi.nlm.nih.gov/nuccore/M73817 ''Clostridium cellulovorans'' cellulose binding protein gene (cbp A)] (CBDclos). At that point we decided to use these two domains. Staying within the cellulose binding domain-family and leave other protein domains like carbohydrate binding domains aside will keep the results comparable. Like changing to a different binding material would change the binding capacities of both domains in the same way. Also we would stay within bacterial CBD and didn't have to spend time thinking about post-translational modification and glycosylation. |
| - | To get to know more about these two domains and | + | To get to know more about these two domains, their properties and their proteins we consulted NCBI and used the [http://blast.ncbi.nlm.nih.gov/ BLAST]-tool to identify the cellulose binding domains and ExPASy-tools for further measurements. |
[[Image:Bielefeld2012_pfam00553.jpg|130px|thumb|left|Figure 5: Crystal structure of the CBM_2-family]] | [[Image:Bielefeld2012_pfam00553.jpg|130px|thumb|left|Figure 5: Crystal structure of the CBM_2-family]] | ||
[[Image:Bielefeld2012_cfimiexo.jpg|500px|thumb|right|Figure 6: Protein-BLAST of [http://www.ncbi.nlm.nih.gov/nucleotide/327179207?report=genbank&log$=nucltop&blast_rank=3&RID=152ZCN0E01N ''Cellulomonas fimi'' ATCC 484 exoglucanase gene]]] | [[Image:Bielefeld2012_cfimiexo.jpg|500px|thumb|right|Figure 6: Protein-BLAST of [http://www.ncbi.nlm.nih.gov/nucleotide/327179207?report=genbank&log$=nucltop&blast_rank=3&RID=152ZCN0E01N ''Cellulomonas fimi'' ATCC 484 exoglucanase gene]]] | ||
| - | The CBD of the Cellulomonas fimi ATCC 484 exoglucanase gene (Figure 6) is a C-terminal | + | The CBD of the Cellulomonas fimi ATCC 484 exoglucanase gene (Figure 6) is a 100 amino acid long domain, close to the C-terminal ending of the protein with a theoretical pI of 8.07 and a molecular weight of 10.3 kDa. It is classified to be stable and belongs to the Cellulose Binding Modul family 2 (pfam00553/cl02709). This means two tryptophan residues are involved in cellulose binding, this type of CBD is only found in bacteria. Also a CBM49 Carbohydrate binding domain is found within the protein domain, where [http://www.ncbi.nlm.nih.gov/pubmed/17322304?dopt=Abstract binding studies] have shown, that it binds to crystalline cellulose, which could be a possible target for immobilization. |
[[Image:Bielefeld2012_ccellubp.jpg|450px|thumb|left|Figure 7: Protein-BLAST of [http://www.ncbi.nlm.nih.gov/nuccore/M73817 ''Clostridium cellulovorans'' cellulose binding protein gene (cbp A)]]] | [[Image:Bielefeld2012_ccellubp.jpg|450px|thumb|left|Figure 7: Protein-BLAST of [http://www.ncbi.nlm.nih.gov/nuccore/M73817 ''Clostridium cellulovorans'' cellulose binding protein gene (cbp A)]]] | ||
[[Image:Bielefeld2012_pfam00942.jpg|130px|thumb|right|Figure 8: Crystal structure of the CBM_3-family]] | [[Image:Bielefeld2012_pfam00942.jpg|130px|thumb|right|Figure 8: Crystal structure of the CBM_3-family]] | ||
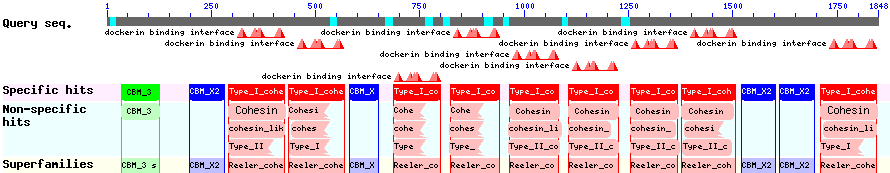
| - | The CBD of the [http://www.ncbi.nlm.nih.gov/nuccore/M73817 ''Clostridium cellulovorans'' cellulose binding protein gene (cbp A)] on the other hand is a N-terminal | + | The CBD of the [http://www.ncbi.nlm.nih.gov/nuccore/M73817 ''Clostridium cellulovorans'' cellulose binding protein gene (cbp A)] on the other hand is a N-terminal domain with 92 amino acids, theoretical pI of 4.56 and ist also classified as stable. It belongs to the Cellulose Binding Modul family 3 (pfam00942/cl03026) and is part of a very large cellulose binding protein with four other carbohydrate binding moduls and a lot of docking interfaces for the proteins in its amino acid sequence (Figure 7). |
| Line 34: | Line 34: | ||
[[image:Bielefeld2012_GFP.jpg|300px|thumb|right|Aequorea victoria green fluorescent protein in action]] | [[image:Bielefeld2012_GFP.jpg|300px|thumb|right|Aequorea victoria green fluorescent protein in action]] | ||
| - | To measure the capacity and strength of the bonding between the cellulose binding domains and different types of cellulose many different assays have been made. One of the simplest and most often used is the fusion of the CBD to a reporter-protein, especially [http://www.ncbi.nlm.nih.gov/pubmed/ | + | To measure the capacity and strength of the bonding between the cellulose binding domains and different types of cellulose many different assays have been made. One of the simplest and most often used is the fusion of the CBD to a reporter-protein, especially [http://www.ncbi.nlm.nih.gov/pubmed/22305911 green or red fluorescent protein (GFP/RFP)] is very common. The place of the CBD is measured through the fluorescence of the fused GFP and quantification can easily been done. |
[http://www.ncbi.nlm.nih.gov/pubmed/18573384 Protocol]: | [http://www.ncbi.nlm.nih.gov/pubmed/18573384 Protocol]: | ||
| Line 49: | Line 49: | ||
==Cloning of the Cellulose Binding Domains== | ==Cloning of the Cellulose Binding Domains== | ||
| - | The cloning of the CBDs should fit to the cloning of our laccases, so we designed the BioBricks with a T7-promoter and the B0034 RBS to have a similar method of cultivation. After we investigated the restriction-sites it showed, that at least for the characterization of the CBDs a quick in-frame assembly of the CBDs and a GFP would be possible, because nether the CBDs nor the GFP (<partinfo>I13522</partinfo>) of the partsregistry inherits a ''Age''I- or ''Ngo''MIV-site, which makes Freiburg-assembly possible. To do so, we designed [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Primers primers] for the constructs <partinfo>BBa_K863101</partinfo> (CBDcex(T7)), carrying the CBDcex domain from the ''C. fimi'' exoglucanase and <partinfo>BBa_K863111</partinfo> (CBDclos(T7)), carrying the CBDclos domain from the ''C.cellulovorans'' binding protein. The protein-BLAST of the two CBDs gave a exact picture of which bases are count to the binding domains and which are not. To be sure not to disturb the folding anyway we kept 6 to 12 bases up and downstream of the domains as conserved sequences. | + | The cloning of the CBDs should fit to the cloning of our laccases, so we designed the BioBricks with a T7-promoter and the B0034 RBS to have a similar method of cultivation. After we investigated the restriction-sites it showed, that at least for the characterization of the CBDs a quick in-frame assembly of the CBDs and a GFP would be possible, because nether the CBDs nor the GFP (<partinfo>I13522</partinfo>) of the partsregistry inherits a ''Age''I- or ''Ngo''MIV-site, which makes Freiburg-assembly possible. To do so, we designed [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Primers primers] for the constructs <partinfo>BBa_K863101</partinfo> (CBDcex(T7)), carrying the CBDcex domain from the ''C. fimi'' exoglucanase and <partinfo>BBa_K863111</partinfo> (CBDclos(T7)), carrying the CBDclos domain from the ''C.cellulovorans'' binding protein. The protein-BLAST of the two CBDs gave a exact picture of which bases are count to the binding domains and which are not. To be sure not to disturb the folding anyway we kept 6 to 12 bases up and downstream of the domains as conserved sequences. Even if the [http://partsregistry.org/Part:BBa_K863101 CBDcex]is a N-terminal domain we chose to make the domains N-terminal, so the BioBrick already carries all regulatory parts and the linked protein can easily be exchanged. This also would be nice for other people using this part. |
<center> | <center> | ||
| Line 65: | Line 65: | ||
The listed complementary primers added, besides the Freiburg-suffix, a two amino acid Glycine-Serine-linker to the end of the CBDs. This is a very short linker, but as GFP-experts and [http://www.ncbi.nlm.nih.gov/pubmed/17394253 publications] described GFP and CBDs are very stable proteins and should cope with a very short linker. The benefit of a short linker is that protease activity is kept minimal. | The listed complementary primers added, besides the Freiburg-suffix, a two amino acid Glycine-Serine-linker to the end of the CBDs. This is a very short linker, but as GFP-experts and [http://www.ncbi.nlm.nih.gov/pubmed/17394253 publications] described GFP and CBDs are very stable proteins and should cope with a very short linker. The benefit of a short linker is that protease activity is kept minimal. | ||
| - | The GFP <partinfo>K863121</partinfo> we wanted to | + | The GFP <partinfo>K863121</partinfo> we wanted to use for the assay was an alternated version of the <partinfo>BBa_I13522</partinfo>. We made [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Primers primers] that added a Freiburg-pre- and suffix to the GFP coding sequence and a His-tag to the C-terminus to get it purified for the measurements. This part <partinfo>BBa_K863121</partinfo> (GFP_His) should be easily added to the CBDs and assemble to the fusion-proteins <partinfo>BBa_K863103</partinfo> CBDcex(T7)+GFP_His] and <partinfo>BBa_K863113</partinfo> CBDclos(T7)+GFP_His]). |
<center> | <center> | ||
| Line 75: | Line 75: | ||
</center> | </center> | ||
| + | Since we added a His-tag to the end of the GFP, we had to make a alternated version of the <partinfo>BBa_I13522</partinfo> to compare binding of GFP with and without CBD. Therefore we made a forward primer, to amplify the whole <partinfo>BBa_I13522</partinfo> and used the GFP_His_compl-primer to add the His-tag to the C-terminus. | ||
| + | |||
| + | <center> | ||
| + | {|class="wikitable" | ||
| + | | GFP_FW_SV || 39 || acgtcacctgcgtgtagctCGTAAAGGAGAAGAACTTTT | ||
| + | |} | ||
| + | </center> | ||
Due to bad cleavage efficiency at the ''Pst''I restiction-site in nearly all PCR-products the cloning of the CBDs ([http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)])and especially the insertion of [http://partsregistry.org/Part:BBa_K863121 GFP_His] to the CBDs took a lot more time as expected. This was because, while designing the primers, we missed to add additional base pairs from the cleavage-site to the termini to increase the efficiency to standard. When we got aware of the problem, cloning got a lot quicker and more successful. | Due to bad cleavage efficiency at the ''Pst''I restiction-site in nearly all PCR-products the cloning of the CBDs ([http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)])and especially the insertion of [http://partsregistry.org/Part:BBa_K863121 GFP_His] to the CBDs took a lot more time as expected. This was because, while designing the primers, we missed to add additional base pairs from the cleavage-site to the termini to increase the efficiency to standard. When we got aware of the problem, cloning got a lot quicker and more successful. | ||
| Line 92: | Line 99: | ||
</center> | </center> | ||
[[File:Bielefeld2012_S3N10linker.jpg|500px|thumb|right|PCR-product of BBa_K863104 with S3N10-primers]] | [[File:Bielefeld2012_S3N10linker.jpg|500px|thumb|right|PCR-product of BBa_K863104 with S3N10-primers]] | ||
| - | Sadly the primers arrived just a few days before wiki freeze and we had no time to test that. The switching to the constitutive promoter had no obvious effect (no green colonies or culture). Which led us to the conclusion, that the problem has to be the | + | Sadly the primers arrived just a few days before wiki freeze and we had no time to test that. The switching to the constitutive promoter had no obvious effect (no green colonies or culture). Changing the expression strain of ''E. coli'' BL21 didn't do no positive effect ether. Which led us to the conclusion, that besides further information the problem has to be the space in between the two proteins and the CBD and GFP must hamper each other from folding correctly. To test and solve this a very long linker with three serines followed by ten asparagines should be assembled in the already existing parts via a blunt end cloning. This also is right at hand, just wasn't successful in the time which was left. |
<center> | <center> | ||
{|class="wikitable" | {|class="wikitable" | ||
| Line 102: | Line 109: | ||
|} | |} | ||
</center> | </center> | ||
| + | == Literature == | ||
| + | Biotechnol Bioeng. 2007 Oct 15;98(3):599-610. | ||
| + | Strategy for selecting and characterizing linker peptides for CBM9-tagged fusion proteins expressed in Escherichia coli. | ||
| + | Kavoosi M, Creagh AL, Kilburn DG, Haynes CA. | ||
| + | J Biol Chem. 2007 Apr 20;282(16):12066-74. Epub 2007 Feb 23. | ||
| + | A tomato endo-beta-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49). | ||
| + | Urbanowicz BR, Catalá C, Irwin D, Wilson DB, Ripoll DR, Rose JK. | ||
| + | Protein Expr Purif. 2012 Apr;82(2):290-6. Epub 2012 Jan 28. | ||
| + | Cellulose affinity purification of fusion proteins tagged with fungal family 1 cellulose-binding domain. | ||
| + | Sugimoto N, Igarashi K, Samejima M. | ||
| + | Anal Chim Acta. 2008 Jul 28;621(2):193-9. Epub 2008 May 27. | ||
| + | Bioseparation of recombinant cellulose-binding module-proteins by affinity adsorption on an ultra-high-capacity cellulosic adsorbent. | ||
| + | Hong J, Ye X, Wang Y, Zhang YH. | ||
Latest revision as of 12:42, 25 September 2012
Contents |
Cellulose Binding Domain - Introduction
In the field of cheap protein-extraction cellulose binding domains (CBD) have made themselves a name. A lot of publications have been made, concerning a cheap strategy to capture a protein from the cell-lysate with a CBD-tag. Also enhanced segregation with CBD-tagged proteins have been observed. Here the idea is different, we want to take advantage of the binding capacity of binding domains not only for purification reasons (it is still a benefit), but also as an immobilizing-protocol for our laccases.
To make a purification and immobilization-tag out of a protein domain, there are a lot of decisions and characterizations you have to get through.
Starting with the choice of the binding domain, the first limitation is accessibility. Our first place to look was, of course the [http://partsregistry.org Partsregistry]. We found a promising Cellulose binding motif of the C. josui Xyn10A gene (<partinfo>BBa_K392014</partinfo>) there and ordered it right from the spot for our project. After some research later concerning the sequence of that BioBrick it turned out that the part is not the CBD of the Xylanase as it should be, but the glycosyl hydrolase domain of the protein (Figure 1). This result made the part useless for our project ([http://partsregistry.org/Part:BBa_K392014:Experience complete review]) and it was the only binding domain in the [http://partsregistry.org Partsregistry] that fitted to our project.
So we started to search the accessible organisms we had via NCBI for binding-domains, -proteins and -motifs and asked in work-groups if they could help us out. The results of our database research were only two chitin/carbohydate binding modules within the Bacillus halodurans genome (we ordered that stain for it's laccase <partinfo>BBa_K863020</partinfo>). One is in the [http://www.ncbi.nlm.nih.gov/nucleotide/289656506?report=genbank&log$=nuclalign&blast_rank=1&RID=0JPT9WMS01N Cochin chitinase gene] and the other in a [http://www.ncbi.nlm.nih.gov/protein/BAB05022.1 chitin binding protein].
Meanwhile the Fermentation group of our university offered use two plasmids (p570 & p671), containing two different cellulose binding domains. The Cellulose binding domain of the [http://www.ncbi.nlm.nih.gov/nucleotide/327179207?report=genbank&log$=nucltop&blast_rank=3&RID=152ZCN0E01N Cellulomonas fimi ATCC 484 exoglucanase gene] (CBDcex) and the Cellulose binding domain of [http://www.ncbi.nlm.nih.gov/nuccore/M73817 Clostridium cellulovorans cellulose binding protein gene (cbp A)] (CBDclos). At that point we decided to use these two domains. Staying within the cellulose binding domain-family and leave other protein domains like carbohydrate binding domains aside will keep the results comparable. Like changing to a different binding material would change the binding capacities of both domains in the same way. Also we would stay within bacterial CBD and didn't have to spend time thinking about post-translational modification and glycosylation.
To get to know more about these two domains, their properties and their proteins we consulted NCBI and used the [http://blast.ncbi.nlm.nih.gov/ BLAST]-tool to identify the cellulose binding domains and ExPASy-tools for further measurements.
The CBD of the Cellulomonas fimi ATCC 484 exoglucanase gene (Figure 6) is a 100 amino acid long domain, close to the C-terminal ending of the protein with a theoretical pI of 8.07 and a molecular weight of 10.3 kDa. It is classified to be stable and belongs to the Cellulose Binding Modul family 2 (pfam00553/cl02709). This means two tryptophan residues are involved in cellulose binding, this type of CBD is only found in bacteria. Also a CBM49 Carbohydrate binding domain is found within the protein domain, where [http://www.ncbi.nlm.nih.gov/pubmed/17322304?dopt=Abstract binding studies] have shown, that it binds to crystalline cellulose, which could be a possible target for immobilization.
The CBD of the [http://www.ncbi.nlm.nih.gov/nuccore/M73817 Clostridium cellulovorans cellulose binding protein gene (cbp A)] on the other hand is a N-terminal domain with 92 amino acids, theoretical pI of 4.56 and ist also classified as stable. It belongs to the Cellulose Binding Modul family 3 (pfam00942/cl03026) and is part of a very large cellulose binding protein with four other carbohydrate binding moduls and a lot of docking interfaces for the proteins in its amino acid sequence (Figure 7).
The Binding Assay
To measure the capacity and strength of the bonding between the cellulose binding domains and different types of cellulose many different assays have been made. One of the simplest and most often used is the fusion of the CBD to a reporter-protein, especially [http://www.ncbi.nlm.nih.gov/pubmed/22305911 green or red fluorescent protein (GFP/RFP)] is very common. The place of the CBD is measured through the fluorescence of the fused GFP and quantification can easily been done.
[http://www.ncbi.nlm.nih.gov/pubmed/18573384 Protocol]:
- Harvest the E coli-cells producing the fusion-protein of CBD and GFP and centifuge 10 minutes at top speed.
- Re-suspend the cell-pellet in 50 mM Tris-HCl-Buffer (pH 8.0).
- Break down cells via sonication.
- Centrifuge at top speed for 20 minutes to get rid of the cell-debris.
- Take the supernatant and measure the emission at 511 nm (excitation at 501 nm) (<partinfo>E0040</partinfo>)
- Mix a definite volume of lysate with a definite volume or mass of e.g. crystalline cellulose (CC) or reactivated amorphous cellulose (RAC)
- Wait 15 (RAC) to 30 (CC) minutes
- Take supernatant and measure the emission at 511 nm again.
- The difference between the first an the second measurement is the relative quantity of what has bound to the cellulose.
Cloning of the Cellulose Binding Domains
The cloning of the CBDs should fit to the cloning of our laccases, so we designed the BioBricks with a T7-promoter and the B0034 RBS to have a similar method of cultivation. After we investigated the restriction-sites it showed, that at least for the characterization of the CBDs a quick in-frame assembly of the CBDs and a GFP would be possible, because nether the CBDs nor the GFP (<partinfo>I13522</partinfo>) of the partsregistry inherits a AgeI- or NgoMIV-site, which makes Freiburg-assembly possible. To do so, we designed primers for the constructs <partinfo>BBa_K863101</partinfo> (CBDcex(T7)), carrying the CBDcex domain from the C. fimi exoglucanase and <partinfo>BBa_K863111</partinfo> (CBDclos(T7)), carrying the CBDclos domain from the C.cellulovorans binding protein. The protein-BLAST of the two CBDs gave a exact picture of which bases are count to the binding domains and which are not. To be sure not to disturb the folding anyway we kept 6 to 12 bases up and downstream of the domains as conserved sequences. Even if the [http://partsregistry.org/Part:BBa_K863101 CBDcex]is a N-terminal domain we chose to make the domains N-terminal, so the BioBrick already carries all regulatory parts and the linked protein can easily be exchanged. This also would be nice for other people using this part.
| CBDcex_T7RBS | 80 | TGAATTCGCGGCCGCTTCTAGAGTAATACGACTCACTATAGGGAAAGAGGAGAAATAATGGGT CCGGCCGGGTGCCAGGT |
| CBDcex_2AS-Link_compl | 56 | CTGCAGCGGCCGCTACTAGTATTAACCGGTGCTGCCGCCGACCGTGCAGGGCGTGC |
| CBDclos_T7RBS | 73 | CCGCTTCTAGAGTAATACGACTCACTATAGGGAAAGAGGAGAAATAATGTCAGTTGAATTTTACAACTCTAAC |
| CBDclos_2ASlink_compl | 63 | CTGCAGCGGCCGCTACTAGTATTAACCGGTGCTGCCTGCAAATCCAAATTCAACATATGTATC |
The listed complementary primers added, besides the Freiburg-suffix, a two amino acid Glycine-Serine-linker to the end of the CBDs. This is a very short linker, but as GFP-experts and [http://www.ncbi.nlm.nih.gov/pubmed/17394253 publications] described GFP and CBDs are very stable proteins and should cope with a very short linker. The benefit of a short linker is that protease activity is kept minimal.
The GFP <partinfo>K863121</partinfo> we wanted to use for the assay was an alternated version of the <partinfo>BBa_I13522</partinfo>. We made primers that added a Freiburg-pre- and suffix to the GFP coding sequence and a His-tag to the C-terminus to get it purified for the measurements. This part <partinfo>BBa_K863121</partinfo> (GFP_His) should be easily added to the CBDs and assemble to the fusion-proteins <partinfo>BBa_K863103</partinfo> CBDcex(T7)+GFP_His] and <partinfo>BBa_K863113</partinfo> CBDclos(T7)+GFP_His]).
| GFP_Frei | 54 | TACGGAATTCGCGGCCGCTTCTAGATGGCCGGCATGCGTAAAGGAGAAGAACTT |
| GFP_His6_compl | 74 | CTGCAGCGGCCGCTACTAGTATTAACCGGTGTGATGGTGATGGTGATGTTTGTATAGTTCATCCATGCCATGTG |
Since we added a His-tag to the end of the GFP, we had to make a alternated version of the <partinfo>BBa_I13522</partinfo> to compare binding of GFP with and without CBD. Therefore we made a forward primer, to amplify the whole <partinfo>BBa_I13522</partinfo> and used the GFP_His_compl-primer to add the His-tag to the C-terminus.
| GFP_FW_SV | 39 | acgtcacctgcgtgtagctCGTAAAGGAGAAGAACTTTT |
Due to bad cleavage efficiency at the PstI restiction-site in nearly all PCR-products the cloning of the CBDs ([http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)])and especially the insertion of [http://partsregistry.org/Part:BBa_K863121 GFP_His] to the CBDs took a lot more time as expected. This was because, while designing the primers, we missed to add additional base pairs from the cleavage-site to the termini to increase the efficiency to standard. When we got aware of the problem, cloning got a lot quicker and more successful.
As the project went on and the T7-constructs didn't seem to work one suspected reason was that a stop codon (TAA) that accidentally resulted between the RBS and ATG could be the reason for that. To solve the problem we started to change to a constitutive promoter (<partinfo>BBa_J61101</partinfo>) using the Freiburg-assembly. Therefore Freiburg forward primers for the CBDs were made.
| CBDcex_Freiburg-Prefix | 54 | GCTAGAATTCGCGGCCGCTTCTAGATGGCCGGCGGTCCGGCCGGGTGCCAGGTG |
| CBDclos_Freiburg-Prefix | 57 | GCTAGAATTCGCGGCCGCTTCTAGATGGCCGGCTCATCAATGTCAGTTGAATTTTAC |
The second advantage of these primers were, that the order of the fusion proteins could easily been changed, when a Freiburg suffix-primer for the GFP would be available, so we ordered that.
| GFP_Freiburg_compl | 61 | ACGTCTGCAGCGGCCGCTACTAGTATTAACCGGTTTTGTATAGTTCATCCATGCCATGTGT |
Sadly the primers arrived just a few days before wiki freeze and we had no time to test that. The switching to the constitutive promoter had no obvious effect (no green colonies or culture). Changing the expression strain of E. coli BL21 didn't do no positive effect ether. Which led us to the conclusion, that besides further information the problem has to be the space in between the two proteins and the CBD and GFP must hamper each other from folding correctly. To test and solve this a very long linker with three serines followed by ten asparagines should be assembled in the already existing parts via a blunt end cloning. This also is right at hand, just wasn't successful in the time which was left.
| S3N10_Cex_compl | 40 | TTGTTGTTGTTCGAGCTCGAGCCGACCGTGCAGGGCGTGC |
| S3N10_Clos_compl | 40 | TTGTTGTTGTTCGAGCTCGAGCTGCCGCCGACCGTGCAGG |
| S3N10_GFP | 40 | CAATAACAATAACAACAACCGTAAAGGAGAAGAACTTTTC |
Literature
Biotechnol Bioeng. 2007 Oct 15;98(3):599-610. Strategy for selecting and characterizing linker peptides for CBM9-tagged fusion proteins expressed in Escherichia coli. Kavoosi M, Creagh AL, Kilburn DG, Haynes CA. J Biol Chem. 2007 Apr 20;282(16):12066-74. Epub 2007 Feb 23. A tomato endo-beta-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49). Urbanowicz BR, Catalá C, Irwin D, Wilson DB, Ripoll DR, Rose JK. Protein Expr Purif. 2012 Apr;82(2):290-6. Epub 2012 Jan 28. Cellulose affinity purification of fusion proteins tagged with fungal family 1 cellulose-binding domain. Sugimoto N, Igarashi K, Samejima M. Anal Chim Acta. 2008 Jul 28;621(2):193-9. Epub 2008 May 27. Bioseparation of recombinant cellulose-binding module-proteins by affinity adsorption on an ultra-high-capacity cellulosic adsorbent.
Hong J, Ye X, Wang Y, Zhang YH. "
"