Team:LMU-Munich/Bacillus BioBricks/Sporovector
From 2012.igem.org
| (2 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<p align="justify"> | <p align="justify"> | ||
| - | Our '''Sporo'''vector is specially designed to give you the opportunity to easily create Sporobeads with any | + | Our '''Sporo'''vector is specially designed to give you the opportunity to easily create Sporobeads with any protein you can think of. The part between the ''EcoR''I and ''Pst''I is available as BioBrick in pSB1C3 and can be cloned in any of our empty ''B. subtilis'' vectors. We also offer it “precloned” in pSB<sub>''Bs''</sub>4S, which in this version lacks the ''NgoM''IV restriction sites. |
| - | [[File:Sporovectorcloning.png|600px]] | + | </p> |
| - | The '''Sporo'''vector was created by ligation of three cut PCR-products. It is designed to allow N-terminal | + | [[File:Sporovectorcloning.png|600px|center]] |
| - | + | <p align="justify"> | |
| - | + | The '''Sporo'''vector was created by ligation of three cut PCR-products with special overhangs (P<sub>''cotYZ''</sub>, ''mRFP'' and ''cgeA''-B0014). It is designed to allow N-terminal fusions of genes in Freiburg standard ([http://dspace.mit.edu/bitstream/handle/1721.1/45140/BBF_RFC%2025.pdf?sequence=1 RCF25]). Therefore, the gene to be inserted must be cut with ''Xba''I and ''Age''I and the vector must be digested with ''Xba''I and ''NgoM''IV. By this digest, the RFP cassette is removed from this vector to allow easy selection for the integration of the new insert. After ligation, there is a scar of six nucleotides between both genes but no restriction site left. </p> | |
| + | <p align="justify"> | ||
| + | Expression of the gene fusion is controlled by the P<sub>''cot''YZ</sub>-promoter which was the strongest one in our promoter evaluations. The C-terminal gene part is ''cge''A, one of the two known spore crust proteins. | ||
</p> | </p> | ||
<p align="justify"> | <p align="justify"> | ||
Latest revision as of 00:23, 27 September 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

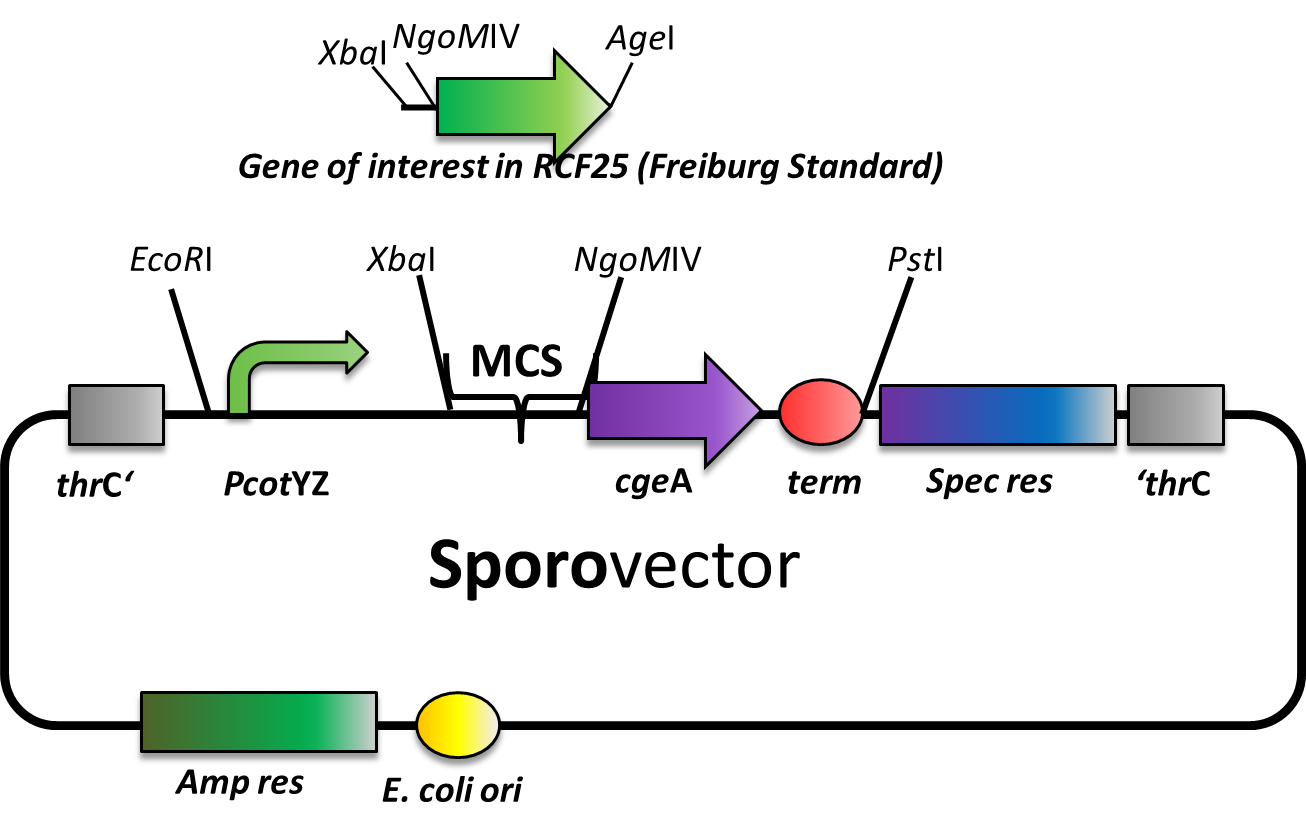
Sporovector
Our Sporovector is specially designed to give you the opportunity to easily create Sporobeads with any protein you can think of. The part between the EcoRI and PstI is available as BioBrick in pSB1C3 and can be cloned in any of our empty B. subtilis vectors. We also offer it “precloned” in pSBBs4S, which in this version lacks the NgoMIV restriction sites.
The Sporovector was created by ligation of three cut PCR-products with special overhangs (PcotYZ, mRFP and cgeA-B0014). It is designed to allow N-terminal fusions of genes in Freiburg standard ([http://dspace.mit.edu/bitstream/handle/1721.1/45140/BBF_RFC%2025.pdf?sequence=1 RCF25]). Therefore, the gene to be inserted must be cut with XbaI and AgeI and the vector must be digested with XbaI and NgoMIV. By this digest, the RFP cassette is removed from this vector to allow easy selection for the integration of the new insert. After ligation, there is a scar of six nucleotides between both genes but no restriction site left.
Expression of the gene fusion is controlled by the PcotYZ-promoter which was the strongest one in our promoter evaluations. The C-terminal gene part is cgeA, one of the two known spore crust proteins.
The vector is cloned but has not been tested with an insert, yet. We are working on further Sporovectors to offer also fusion with cotZ as well as C-terminal fusions.
Back to our BacillusBioBrickBox
 "
"