|
|
| (89 intermediate revisions not shown) |
| Line 14: |
Line 14: |
| | == The "SWITCH" Project == | | == The "SWITCH" Project == |
| | | | |
| - | ===Proteins in medical treatments=== | + | ===In a nutshell=== |
| - | In the past decade, proteins have gained increasing attention as therapeutic agents. Among them, monoclonal antibodies have gained a particular importance, since they present several advantages compared to standard drugs. Reduced risk of non-specific action, longer therapeutic effect and smaller risk of toxicity upon elimination are just several advantages monoclonal antibodies possess when used as drugs.
| + | [[File:File-Team-EPF-Lausanne_main_BD.png|left|frameless|550px]] |
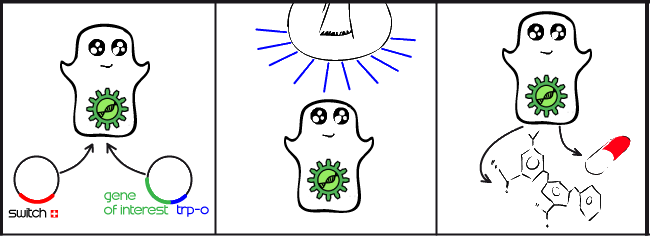
| | + | Our project is all about implementing a light activated genetic switch in mammalian cells. The production of complex proteins is becoming an increasingly commonplace task, particularly in the pharmaceutical industry. Mammalian cells are an ideal environment to make these proteins, but producing them at the right time and optimal rate requires an expression system controlled by a specific signal. Until now, these systems have mostly been limited to chemical signals which have many drawbacks. By using a light-activated expression system, we hope to enable mammalian cells to respond to a light signal, rather than a chemical signal, by initiating gene expression for a protein of interest. |
| | | | |
| - | ===Problems with synthesis=== | + | ===A new expression system: LovTAP fusion protein=== |
| - | Despite all of their advantages, monoclonal antibodies are sometimes challenging to produce. The majority of them are manufactured by recombinant cells that are cultured in huge volumes (called “bioreactors”). In order to achieve a reasonable price for the final product, the antibody needs to be produced at a high concentration. Unfortunately, many antibodies are toxic to the producing cells at high concentrations, which means they get killed before a sufficient concentration of final product is reached.
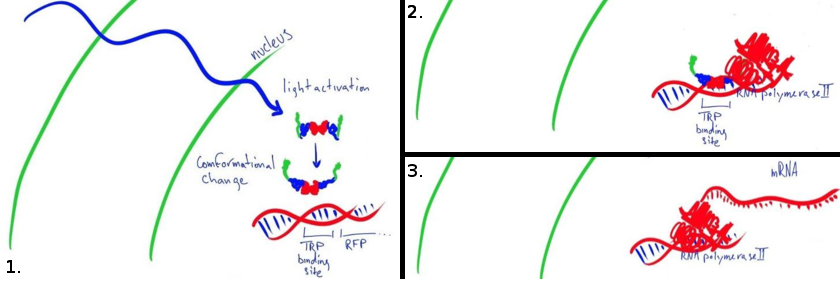
| + | [[File:Team-EPF-Lausanne_activation_alt.png|center|thumb|650px| LovTAP-VP16 gets activated by blue light (1.), then binds to the DNA (2.) and activates the transcription (3.)]] |
| | | | |
| - | So far, this problem has been solved by using cells that are designed to produce the desired protein only when they uptake and sense a particular small molecule. With this technique, cells begin by growing normally and start to produce the desired protein only when the small molecule signal is present in sufficient concentration to trigger the production of the desired product at an optimal concentration, maintaining a balance between protein production and cell health. Unfortunately, this method has a couple of disadvantages. First, the “switch” molecule is often difficult and costly to remove. Second, redesigning cells so that they react to the “switch” molecule can perturb many biological pathways, which in turn decreases the efficiency of antibody production by disrupting normal cell growth and proliferation.
| |
| | | | |
| - | ===Cool solution===
| |
| - | We thought that the best solution for this problem would be a simple switch, that would activate the synthesis of a therapeutic protein in cells just by shining a specific wavelength of light on the cell culture. Our hope was that this method would have a minimal effect on cellular pathways and reduce the effect on cell health. This way the cells would grow happily in darkness and start producing the toxic protein only when light is supplied. With such a switch, there would be nothing to remove from the cell culture, allowing recombinant proteins to be produced more economically than by supplying large amounts of a small molecule trigger.
| |
| | | | |
| - | ===How are we going to do it: LovTAP protein===
| + | Our main goal this summer was to implement a previously untested expression system in mammalian cells. The LovTAP-VP16 fusion protein we used is based on a previously tested fusion protein, [http://partsregistry.org/wiki/index.php/Part:BBa_K191006 LovTAP] by the [https://2009.igem.org/Team:EPF-Lausanne 2009 EPF Lausanne team], which binds to a specific sequence of DNA once it is activated with blue light. By attaching an activating domain to the protein that is known to work in mammalian cells, we hoped to enable LovTAP-VP16 to bind DNA after exposure to light and regulate expression of a gene next to its binding site in a mammalian cell line. |
| - | A really nice way to realize our idea would be to use the LovTAP protein. When illuminated with a blue light, LovTAP changes configuration and acts as a negative regulator of gene expression in bacteria, allowing to switch on and off protein production only by the light. Unfortunately, LovTAP does nothing in the mammalian cells, which are the ones used for the synthesis of the antibodies. We've decided to overcome this problem by attaching a VP16 viral promoter domain to the LovTAP protein, so that it will become a powerful positive regulator of gene expression in mammalian cells. If we transect the LovTAP-VP16 construct along with the gene of the protein we would like to synthesize, we could use it as the perfect light “switch”. It will turn on the protein production when some light will be shined on the cell and won't disturb any pathways.
| + | |
| | | | |
| | ===Another approach: Melanopsin=== | | ===Another approach: Melanopsin=== |
| - | An another nice tool to realize a light-induced switch would be to use a light receptor that already exists in mammalian cells, for instance melanopsin. This way, the receptor will sense the light and transmit a chemical signal to the rest of the cell in order to activate the production of the desired protein. Even though this switch is more complex then the previous one and will affect several pathways, it still eliminates the need for an activating molecule. In addition, it might be more efficient then the LovTAP-VP16 based switch, since it uses a protein that already exists in mammalian cells, and not a fusion protein.
| + | [[File:Team-EPF-Lausanne-melanopsinpathway.JPG|thumb|left|250px| |
| | + | [[Team:EPF-Lausanne/References#Fussenegger_2011|Fusseneger 2011 et al.]] ]] |
| | + | We also worked with another light-activated expression system that has already been proven to work. Martin Fusseneger et al. have designed a pathway that makes extensive use of gene expression pathways already present in the cell. By adding a light-activated receptor on the cell membrane, they promote the release of calcium into the cytoplasm upon exposure to light. This activates the native cellular response to an increased calcium concentration. Promoter proteins sensitive to calcium will react to this change and start expressing genes with specific promoter sites. By introducing a new gene with a calcium-sensitive promoter next to it, we can use light to induce its expression. |
| | | | |
| | ===CHO cells & Co=== | | ===CHO cells & Co=== |
| - | In order to check the validity of our ideas, we attempted to implement both light-induced switches in CHO cells.
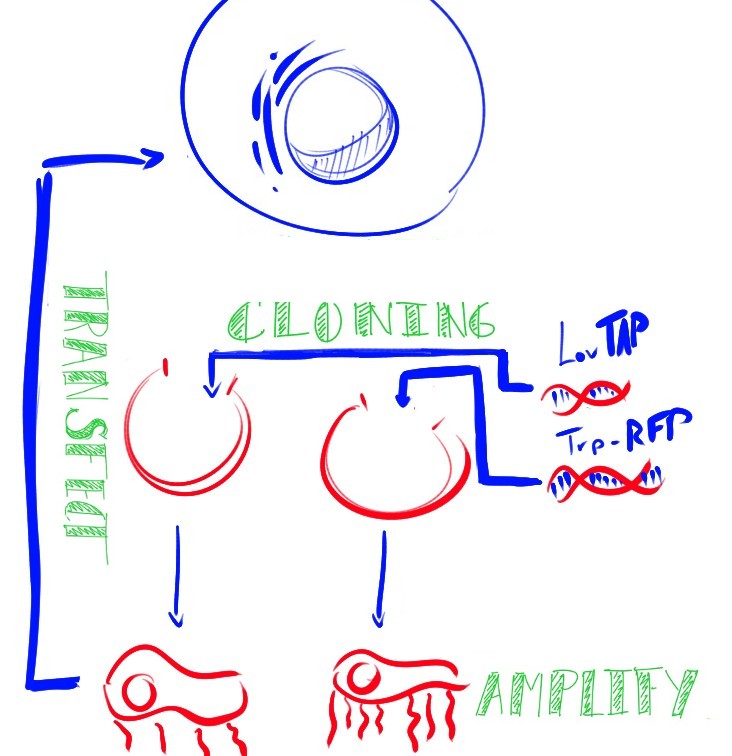
| + | [[File:Team-EPF-Lausanne transfection graphic.jpg |thumb|right|frameless |300px]] |
| - | For the LovTAP-VP16 switch, we've transfected CHO cells with LovTAP-VP16 encoding plasmid along with a reporter plasmid containing a protein that fluoresces in red (dsRed). We can tell that the switch functions correctly if after illumination with blue light we can see a substantial increase in red fluorescence in the cells. | + | CHO (Chinese Hamster Ovary) cells are the workhorses of industrial protein production. The light activated expression systems we worked on would be extremely convenient for production of proteins in an industrial context in these cells. For our project, we transfected these cells with each expression system and conducted a variety of tests. Along with the CHO cell line, we also chose to test these light activated systems using HEK (Human Embryonic Kidney) cells since the melanopsin system had already been shown to work in this cell type. The melanopsin system would provide data to compare to our LovTAP-VP16 system. |
| - | For the Melanopsin - based switch, we will transfect a plasmid expressing a natural light receptor (Melanopsin), along with a reporter that would synthesize the green-fluorescent protein in response to the signaling pathway that is naturally triggered by Melanopsin. We can tell that the switch functions correctly in case we see a substantial increase in green fluorescence in cell culture after illumination with blue light.
| + | |
| | | | |
| | | | |
| - | <!--
| |
| - | ===The Problem===
| |
| - | Producing complex therapeutic proteins requires biosynthesis in mammalian cells. Such proteins can sometimes have a certain level of toxicity for the cells and limit their productivity if they are produced constantly and are accumulating. To avoid this, the pharmaceutical industry use 'rewired' cells that synthesize toxic proteins only when a special molecule is added to the bioreactor. This solution has two disadvantages. First, cell rewiring affects several pathways and decreases cell productivity. Second, the 'special' molecule will mix with the final product and a purification will be needed to get rid of it.
| |
| | | | |
| - | The pharmaceutical industry needs an easier way to induce the production of a specific compound in mammalian cell.
| + | In each different cell line, we tested a transfection containing a light-sensitive protein and a target gene or readout. We chose to express easily detectable fluorescent proteins so that we could compare the efficiency of both systems in turning on a particular gene. The fluorescent protein gene could eventually be replaced by a gene coding for a medically-relevant protein of interest to the pharmaceutical industry. |
| | | | |
| - | This is where our "SWITCH" project steps in.
| + | ===Not Only Pharma=== |
| - | | + | [[File:Team-EPF-Lausanne-very motivated cell dressed as hamster.png|150px|right]] |
| - | ===The Simple Switch=== | + | The use of our switch is not exclusively limited to the pharmaceutical industry. The LovTAP-VP16 switch can also be used in experimental biology to observe temporal gene expression suppression. Our project also has a multitude of other valuable applications, yet achieving them requires comprehension and consent of general public. That's why, in addition to our experimental work, we've presented our project and introduced the public to the field of synthetic biology, and attempted to engage in a two-sided dialog by polling different populations to get their opinion about synthetic biology. |
| - | We want to design a light-sensitive switch, so that the product will only be synthesized by the cells in presence of blue light, allowing them to grow happily in the dark.
| + | |
| - | The switch is based on an already existing chimeric protein, LovTAP. This protein was originally intended to act as a light-induced repressor in bacteria. The EPFL 2009 iGEM team proposed to fuse it with VP16, a viral activator, in order to convert it into a light-induced activator in mammalian cells. This way, it will be enough to add a LovTAP binding site in front of the gene of interest and a LovTAP encoding gene to induce protein expression without using any additional chemical or disturbing too many pathways. | + | |
| - | | + | |
| - | This year we will try to realize this idea by transfecting CHO (Chinese hamster ovary) cells with two plasmids: one coding for the LovTAP-VP16 fusion protein and another with a read-out ( red fluorescence ) protein preceded by a binding site for LovTAP-VP16. If everything goes as expected, LovTAP-VP16 will only bind the plasmid and activate the production of read-out when exposed to light.
| + | |
| - | | + | |
| - | ===The Complex Switch===
| + | |
| - | | + | |
| - | In addition to LovTAP switch, we will be realizing a more complex, melanopsin-based light switch, as described by Fussenegger et al. In this switch, light-sensitive melanopsin triggers a cascade involving calcium ion channels that eventually leads to the transcription of the gene of interest. The switch involves more complex signalling cascades, but it also uses only one natural protein and might function better then the simple switch. Fussenegger's team did this using HEK (human embryo kidney) cells, and we will also try to make it work on CHO cells, so that we can determine which switch works better in the CHO cells.
| + | |
| - | | + | |
| - | -->
| + | |
| | | | |
| | + | Check the rest of our wiki to see our project more in detail! |
| | | | |
| | {{:Team:EPF-Lausanne/Template/Footer}} | | {{:Team:EPF-Lausanne/Template/Footer}} |
 "
"