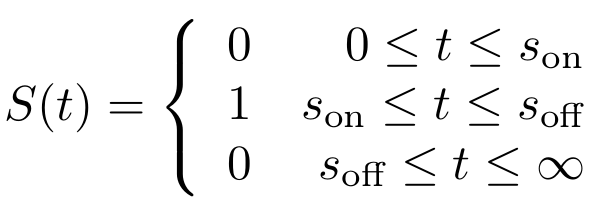Team:Amsterdam/modeling/odemodel
From 2012.igem.org
(Difference between revisions)
(→In practice) |
|||
| (16 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
Using the here presented model, we will examine how to infer the signal detection time from the amount of methylated plasmids. The cellular division rate determines how long a signal is stored in the ''Cellular Logbook''. | Using the here presented model, we will examine how to infer the signal detection time from the amount of methylated plasmids. The cellular division rate determines how long a signal is stored in the ''Cellular Logbook''. | ||
| - | All units are dimensionless in this model, as its sole purpose is clarification of usage of the 'Cellular Logbook'. | + | All units are dimensionless in this model, as its sole purpose is clarification of practical usage of the ''Cellular Logbook''. |
= Inferring the time of signal onset = | = Inferring the time of signal onset = | ||
| Line 14: | Line 14: | ||
== Methylated bits over time == | == Methylated bits over time == | ||
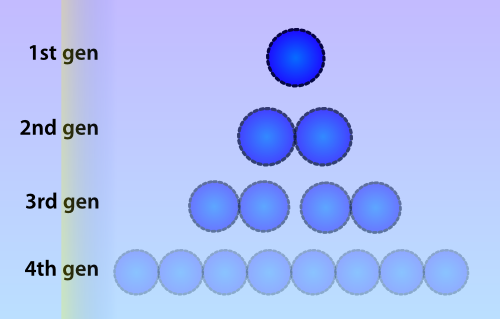
| - | Numerous | + | Numerous identical plasmids are often present in single cells and plasmids replicate independently of the bacterial chromosome (Scott 1984). A plasmid copy number (PCN) has been determined for all plasmids in the Parts Registry, which indicates a likely amount of copies of the plasmid to be present in each cell. Unlike eukaryotes, prokaryotes do not copy DNA methylation patterns to the newly synthesized strand during DNA replication. This will lead to a dilution of the amount of ‘written’-plasmids over time, mostly due to cell replication and the ensuing binomial division of the plasmids in the parent cell among the two daughter cells. |
| - | [[File:Celldivision.png|thumb|300px|Due to cell division, the amount of methylated plasmids will be approximately halved during each | + | [[File:Celldivision.png|thumb|300px|Due to cell division, the amount of methylated plasmids will be approximately halved during each division cycle. In this picture a lower opacity indicates a lower amount of methylated plasmids]] |
The volatilty of this memory design seemed a downside at first, but quickly opened our eyes to a very exciting feature of this system. By analyzing the fraction: | The volatilty of this memory design seemed a downside at first, but quickly opened our eyes to a very exciting feature of this system. By analyzing the fraction: | ||
| Line 22: | Line 22: | ||
<math>F(t) = \frac{\text{written plasmids}}{\text{written + unwritten plasmids}}</math> | <math>F(t) = \frac{\text{written plasmids}}{\text{written + unwritten plasmids}}</math> | ||
</center> | </center> | ||
| - | at the time of memory read-out, the time at which the signal was registered | + | at the time of memory read-out, the time at which the signal was registered can be inferred. |
== Model definition == | == Model definition == | ||
| + | First, let’s model the input signal/compound which is to be reported on. Imagine the to be stationary and positioned along a fluidic stream so that the signal to be registered can pass the ''Cellular Logbook''. Modelling the signal using the piecewise function <math>S(t)</math> now seems appropriate. Here, <math>s_{\text{on}}</math> is defined as the time at which the signal is first encountered and <math>s_{\text{off}}</math> as the time at which the signal is turned off. | ||
| - | + | [[File:Signalformula.png|center|frameless|200px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
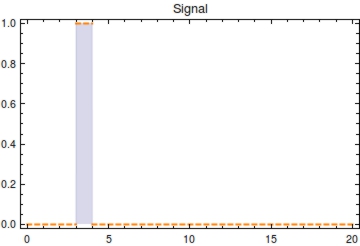
[[File:Signal.jpg|image|thumb|300px|Plot of the input signal <math>S(t)</math> with <math>s_{\text{on}}</math> at 3 and <math>s_{{\text{off}}}</math> at 4]] | [[File:Signal.jpg|image|thumb|300px|Plot of the input signal <math>S(t)</math> with <math>s_{\text{on}}</math> at 3 and <math>s_{{\text{off}}}</math> at 4]] | ||
| - | |||
We will model a single cell with multiple identical plasmids. Each of the plasmid copies contain the gene for the methyltransferase and the so called ''bit region'', which is the region especially purposed to be methylated in presence of a signal. The following assumptions/conditions are made: | We will model a single cell with multiple identical plasmids. Each of the plasmid copies contain the gene for the methyltransferase and the so called ''bit region'', which is the region especially purposed to be methylated in presence of a signal. The following assumptions/conditions are made: | ||
| Line 84: | Line 74: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | |||
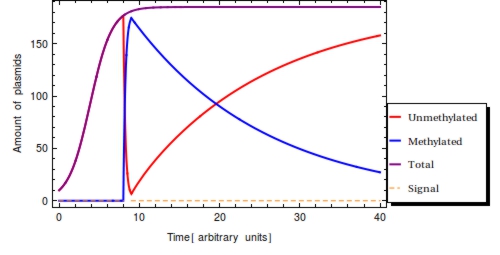
| + | Using the parameter values of Table 1 a simulation with a duration of 40 time units is shown in Figure 2. The plasmid population within a ''Cellular Logbook'' is shown to be completely converted to methylated plasmids shortly after <math>s_{\text{on}}</math>. As long as the signal is still present – until <math>s_{\text{off}}</math>, – the bit on all newly copied plasmids will be immediately methylated as the signal is still present. After <math>s_{\text{off}}</math>, <math>F(t)</math> will start to decrease. This is mostly due to cell division, during which the cell’s plasmids will be binomially distributed between the two two daughter cells, halving the plasmid amount every division cycle. In this simulation, this degradation due to cell division has been accounted for in the constant degradation rate <math>\alpha</math>. The duration of time after which a small trail of methylated plasmids is still present is related to two factors: positively to the amount of methylated cells at <math>s_{\text{off}}</math> and negatively to the plasmid degradation rate. | ||
<table align="right"> | <table align="right"> | ||
| Line 102: | Line 94: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| - | |||
To reinforce that: | To reinforce that: | ||
| Line 131: | Line 120: | ||
The monotonically decreasing value of <math>F(t) = \frac{\text{methylated plasmids}}{\text{total plasmids}}</math> can be used to infer <math>s_{\text{off}}</math>, given that the degradation rate (<math>\alpha</math>) and capacity constraint <math>Ca</math> are known and constant. Also assumed is that all bits are methylated during signal presence, this implicates <math>\omega</math> is sufficient to methylate all bits during presence of the signal. Irrespective of the initial amount of plasmids, the population of plasmids within the single cell will have reached a steady state value of <math>\frac{\beta}{\alpha}</math>. As we see in the Figure 2, <math>F(t)</math> will start to decrease as a function of the degradation rate after the signal has left the medium following the following function: | The monotonically decreasing value of <math>F(t) = \frac{\text{methylated plasmids}}{\text{total plasmids}}</math> can be used to infer <math>s_{\text{off}}</math>, given that the degradation rate (<math>\alpha</math>) and capacity constraint <math>Ca</math> are known and constant. Also assumed is that all bits are methylated during signal presence, this implicates <math>\omega</math> is sufficient to methylate all bits during presence of the signal. Irrespective of the initial amount of plasmids, the population of plasmids within the single cell will have reached a steady state value of <math>\frac{\beta}{\alpha}</math>. As we see in the Figure 2, <math>F(t)</math> will start to decrease as a function of the degradation rate after the signal has left the medium following the following function: | ||
| - | + | $$ | |
| + | \frac{dP_{1}}{dt} = - \alpha\ P_{1} | ||
| + | $$ | ||
| - | Integrating this differential equation | + | Integrating this differential equation and multiplying by the steady value <math>\frac{\beta}{\alpha}</math> will yield the amount of methylated plasmids at time <math>t</math>, given that there were <math>\frac{\beta}{\alpha}</math> methylated plasmids at <math>t = 0</math>. |
| - | + | $$ | |
| - | + | P_{1}(t) = \frac{\beta}{\alpha} e^{-\alpha t} = \frac{\beta}{\alpha} F(t) | |
| - | + | $$ | |
| - | + | ||
| - | + | ||
| - | + | ||
By solving the previous equation, we can calculate the time <math>t</math> that has passed after <math>s_{\text{off}}</math> from <math>F(t)</math>: | By solving the previous equation, we can calculate the time <math>t</math> that has passed after <math>s_{\text{off}}</math> from <math>F(t)</math>: | ||
| - | + | $$ | |
| + | t = \frac{\ln(F(t))}{-\alpha} | ||
| + | $$ | ||
== In practice == | == In practice == | ||
| - | |||
[[File:bands.jpeg|center|thumb|500px|Gel representations for a range of different <math>F(t)</math> values. Complete methylation of all bits results in a single, bright band at the top of the gel. This indicates the undigested, linearized plasmid. Decreasing the amount of methylated bits shifts the intensity of the top band away to the two bottom bands. These indicate the linearized & successfully digested plasmid]] | [[File:bands.jpeg|center|thumb|500px|Gel representations for a range of different <math>F(t)</math> values. Complete methylation of all bits results in a single, bright band at the top of the gel. This indicates the undigested, linearized plasmid. Decreasing the amount of methylated bits shifts the intensity of the top band away to the two bottom bands. These indicate the linearized & successfully digested plasmid]] | ||
In a typical laboratory situation, doing a restriction enzyme assay on the miniprep-extracted plasmid DNA out of followed by gel electrophoresis will be the most convenient way to assess the methylation status of the bits. The relative intensities of the gel bands can then be used to infer <math>F(t)</math>. Unmethylated bits will result in successfully digested DNA fragments and thus two bands of shorter DNA fragments. Methylated bits will not be cut and will therefore result in one longer band, shown more to the top of the gel. Thus the top and two bottom gel bands are mutually exclusive as they indicate the same (linearized) plasmid DNA to either be digested, resulting in the two bottom bands, or undigested, resulting in the top band. A high value for <math>F(t)</math> indicates recent detection of the signal, whereas a low value indicates detection to have occurred longer ago. | In a typical laboratory situation, doing a restriction enzyme assay on the miniprep-extracted plasmid DNA out of followed by gel electrophoresis will be the most convenient way to assess the methylation status of the bits. The relative intensities of the gel bands can then be used to infer <math>F(t)</math>. Unmethylated bits will result in successfully digested DNA fragments and thus two bands of shorter DNA fragments. Methylated bits will not be cut and will therefore result in one longer band, shown more to the top of the gel. Thus the top and two bottom gel bands are mutually exclusive as they indicate the same (linearized) plasmid DNA to either be digested, resulting in the two bottom bands, or undigested, resulting in the top band. A high value for <math>F(t)</math> indicates recent detection of the signal, whereas a low value indicates detection to have occurred longer ago. | ||
| - | To get a hands-on feel of the effects that the plasmid degradation and replication rate have on <math>F(t)</math>, an interactive version in Mathematica is hosted on [https://www.dropbox.com/s/ | + | To get a hands-on feel of the effects that the plasmid degradation and replication rate have on <math>F(t)</math>, an interactive version in Mathematica is hosted on [https://www.dropbox.com/s/2b32sys01ywdotl/timeinferrance.nb Dropbox]. |
| + | This file contains the code for all analyses and graphics (except for the cell division scheme) on this page. | ||
</div> | </div> | ||
Latest revision as of 07:16, 24 September 2012
 "
"