Team:Chalmers-Gothenburg/Notebook
From 2012.igem.org
(→Week 2: 11th - 15th of June) |
m |
||
| (171 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Chalmers-Gothenburg/Template2}} | {{Team:Chalmers-Gothenburg/Template2}} | ||
| - | = | + | <html> |
| + | <head> | ||
| + | <style type="text/css"> | ||
| + | div.box2 | ||
| + | { | ||
| + | width:250px; | ||
| + | height:600px; | ||
| + | background-color:#FFFFFF; | ||
| + | border:0px solid #D8D8D8; | ||
| + | } | ||
| - | + | div.box2 p | |
| - | + | { | |
| + | margin:20px 20px; | ||
| + | font-weight:normal; | ||
| + | color:#000000; | ||
| + | align:justify | ||
| + | } | ||
| - | + | </style> | |
| + | </head> | ||
| - | + | <body> | |
| - | + | <div class="box2"; style="position:absolute;left:700px;top:330px;"> | |
| + | <p><div align="left" style="margin-left:10px; margin-right:10px; "> <b><font size="9"><b>Sponsors</b></font></b> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <a href="http://microsynth.ch/" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/74/Microsynth_Logo_color.jpg" width="220" alt="Microsynth" /></a> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <a href="http://www.gatc-biotech.com/en/index.html" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/c/c1/GATC.gif" width="220" alt="GATC Biotech" /></a> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <a href="http://www.chalmers.se/en/Pages/default.aspx" target="_blank"> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/d/dc/Chalmers40.png" width="150" alt="Chalmers" /></center></a> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <a href="http://www.molnlycke.com/com/" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/6/69/Molnlycke.jpg" width="220" alt="Mölnlycke Health Care" /></a> | ||
| - | + | ||
| + | </p> | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </body> | ||
| + | |||
| + | </html> | ||
| + | |||
| + | |||
| + | <div align="justify" style="width:auto; margin-left:0px; margin-right:290px;"> | ||
| + | __NOEDITSECTION__ | ||
| + | =='''Week 13 - Week 17, 27th of August - 26th of September'''== | ||
| + | |||
| + | During these weeks, we were very busy with paper work, such as the summary, the analysis and discussion of our results and the update of the wiki page. We also started to prepare the presentation of our project for the Jamboree. | ||
| + | |||
| + | In addition, we continued with the cloning of our parts into BioBricks and we managed finally to send in the FIG1 promoter. Another task was the preparation of glycerol stocks of our yeast strains and lastly we had to clean the lab. | ||
| + | |||
| + | SEE YOU IN AMSTERDAM! | ||
| + | |||
| + | =='''Week 12, 20th - 24th of August'''== | ||
| + | '''BioBrick construction''' | ||
| + | |||
| + | We spent most of the time on Miniprep and gelelectrophoresis in order to prepare and verify our BioBrick vectors containing our parts. But the results on the gels were not satisfying so we redid the ligation and transformation once more. Next week we hopefully can finish our BioBricks and send it to the Registry. | ||
| + | |||
| + | '''Indigo assay''' | ||
| + | |||
| + | We decided to do one last experiment with our strains containing the indigo genes. The goal of this experiment was to investigate which compounds that are present in these colourful strains. The strain was grown in shake flasks in both galactose and glucose and after 24h, 2g/l of tryptophan was added. After another 24h, the same redbrown respectively greenbrown colonies as in week 10 were observed. We centrifuged the cells and measured the absorbance spectrum of the colorful supernatant. Some peaks could be observed and next week, we will analyze the spectrum in more detail och hopefully be able to explain the arising colours. | ||
| + | |||
| + | The rest of the time we used for paper work such as summarizing our results, preparing the presentation for the Jamboree and designing the poster. | ||
| + | |||
| + | =='''Week 11, 13th - 17th of August'''== | ||
| + | '''Final hCG test''' | ||
| + | |||
| + | One final hCG test was performed. This time the whole system i.e. the yeast strain with the deleted CWP2 gene and containing both the LHCG-receptor and the indigo genes (with the monooxygenase expressed constitutively and the tryptophanase coupled with the pheromone pathway through the FIG1 promoter) was tested. The strain was grown in SD –His –Ura medium with additional tryptophan (2g/l) and hCG hormone was added. | ||
| + | |||
| + | If the system would work, the hormone should activate the pheromone pathway which in turn should activate the expression of tryptophanase. This would then result in digestion of tryptophan and the same brown colour, which has been observed in the strains containing the activated indigo genes, should appear. Unfortunately, no colour change could be observed. That means that the whole system is not working properly. This could be due the limited permeability of the cell wall or a malfunctioning receptor. | ||
| + | |||
| + | '''BioBricks''' | ||
| + | |||
| + | Besides testing our yeast strain, the task of constructing our BioBricks vectors was time-consuming this week. After another unsuccessful transformation, we discovered that we had used the wrong selection antibiotic. Haha, no wonder that no single colony could be observed! Well, we prepared new plates, this time with chloramphenicol, and repeated the digestion/ligation/transformation. And finally, we got some colonies! Next week, we will purify the plasmids and hopefully send it to the Registry of Standard Biological Parts. | ||
| + | |||
| + | '''Human practices''' | ||
| + | |||
| + | A big event of this week was the presentation about Biotechnology and our project for secondary school and high school students. They were participating in a summer research course. It was a real challenge to explain what we are doing for students that barely know what DNA is. But it was fun and we think the students really liked our presentation. | ||
| + | |||
| + | =='''Week 10, 6th - 10th of August'''== | ||
| + | <!--[[File:Imfd70 hcg.jpg|thumb|200px|Microscopic pictures of the IMFD-70 strain. Cells in the bottom row have had the hCG hormone added to them while the cells on the top row have not.]]--> | ||
| - | ''' | + | '''Receptor group''' |
| - | + | This week we did a last hCG test in which we screened for fluorescence in the cells that we added the hCG hormone to. This time three other strains were used: IMFD-70, IMFD-72 and IMFD-74 and they had all been transformed with LH/CG receptor. These strains contain different types of Gα proteins and our hopes were that these would enable the yeast cells to better couple the LH/CG receptor with the pheromone pathway. However, when doing the hCG testing procedure no significant difference could be observed when examining the cells in the fluorescence microscope: all cells, no matter if they had had the hormone added to them or not, were displaying a weak fluorescence. As before, we are not sure whether or not our receptor is functioning and actually detecting the hormone or if the fluorescence displayed by the cells that have been treated with hCG is only background noise. | |
| - | + | The last thing that we are going to do now is to add hormone to our completed strain that expresses both the LH/CG receptor and the indigo genes. This is our last shot at testing to see if the receptor is functioning. We will do the test next week and we would really like to see some of that color-change that we previously have observed in the indigo strain but now in response to detecting the hormone! | |
| - | We | + | '''Construction of BioBricks''' |
| + | [[File:Rachel constructing Biobricks.jpg|thumb|200px|Rachel is preparing her 22nd gel.]] | ||
| + | We decided to register three new parts, the LHCGR gene with two different signal peptides and the FIG1 promoter. This week, after receiving the necessary primers from Microsynth AG, we started with the cloning of our parts into the BioBrick vector. | ||
| + | The first step was PCR amplification of the three parts. We have become experienced in PCR conditions during this summer and we succeeded directly with the PCR in the first approach. The fragments were purified from gel and digested with PstI and EcoRI for 30 minutes. The vector was also digested with the same enzymes and DpnI additionally, as described in the protocol for BioBricks construction provided by Registry of Standard Biological Parts. The reaction was terminated by putting the samples on 80° C for 10 minutes. A ligation mix with insert, vector, T4 ligase and ligation buffer was prepared for each of the three parts. As a negative control, water instead of insert was added to the mix. The samples were incubated for 3h in room temperature and after that, E.coli was transformed with the hopefully correctly ligated constructs. | ||
| + | The anticlimax arose when we checked the plates the day after. Nothing has grown! The only positive thing was that nothing has grown on the negative control either. We tried to investigate what could have gone wrong. We know that ligation uses to be tricky but we have been very successful earlier and it was actually the first time we did not succeed with ligation. This time we followed the protocol for the BioBrick construction and this means that we did not purify the DNA after digestion. Instead, we terminated the digestion by heating the samples and then proceeded directly with ligation. Another big difference was the incubation time for ligation. In our previous approaches, we have always ligated over night, this time, we only incubated the ligation mix for 3 hours. | ||
| + | So next week, we will just use our own protocol again. Let’s hope it will work! | ||
| - | =Week | + | =='''Week 9, 30th of July - 3rd of August'''== |
| + | [[File:CompleteSystem.jpg|thumb|200px|The system is complete! Is this the next generation pregnancy test?]] | ||
| - | + | The complete system has now been put together! The IMFD-73 strain that has had the CWP2 gene deleted is now expressing the LH/CG receptor and has been transformed with both genes needed for indigo production, one of them being placed under the control of the pheromone-induced FIG1 promoter. Will this system be able to function as a pregnancy test? This is a question that we still need to answer. This week, we therefore continued to test our receptor by adding hormone to our yeast cells. | |
| + | '''Receptor group''' | ||
| + | |||
| + | In contrast with last week, we grew our cells in room temperature before adding hCG. This time we also used the ΔCWP2 strain, which we didn’t treat with any cell wall degrading enzymes before the addition of the hormone. Sadly, approximately the same observations as last week could be made. When examining our cells in the fluorescence microscope, all cells displayed a weak fluorescence, both cells that had been treated with the hormone and cells that had not. | ||
| + | |||
| + | In parallel to the hCG tests that have been carried out, we have now also transformed three other strains with the LH/CG receptor. Next week, these strains will be used in our hCG assay instead and we really hope that these will show some more promising results. | ||
| + | |||
| + | =='''Week 8, 23rd - 27th of July'''== | ||
| + | This week, we focused on investigating our yeast strains that had been transformed with different plasmids. Different assays were performed as described below. | ||
'''Receptor group''' | '''Receptor group''' | ||
| - | This week | + | This week was very exciting but also a bit nerve-racking for us in the receptor group since this was the week when we actually got to test our receptor. On Tuesday, we degraded the cell wall of the IMFD-73 strain that had been transformed with the yLHCGR plasmid and the hLHCGR plasmid. As a control, the strain transformed with the empty pSP-GM1 plasmid was also treated. The hCG hormone was then added to the cells. |
| - | + | The IMFD-73 strain that we have used to express our receptor in, has a GFP gene placed under the control of the FIG1 promoter. So, if the receptor is functioning, the hCG hormone should be able to activate the yeast pheromone pathway and thus induce the expression of the GFP gene. Therefore, we used a fluorescence microscope to examine our cells. To our great disappointment, the level of fluorescence was approximately the same when comparing cells that had been treated with the hormone and cells that had not been treated with the hormone. We were already aware of the risk of having levels of background signaling in our IMFD-73 strain and since the fluorescence intensity was approximately the same in the hormone-treated and the non-treated cells, chances are that our receptor is not functioning and is not producing any output signal. | |
| + | |||
| + | There is still hope though! Next week we will try to grow our yeast in room temperature to hopefully give the cells better chances to express and properly fold the receptor. We will also try to express our receptor in three other strains that also have been optimized for expression of mammalian GPCRs but that are a bit different from the IMFD-73 strain that we have used so far. | ||
| - | + | '''Indigo group''' | |
| + | This week was all about the enzymatic assay on tryptophanase. In total there were three of these performed this week and the results were not that satisfying. The readings of the first one did not show any reliable results on consumption of tryptophan. The second one ended up with a too low concentration of proteins in the cell free extract, so no OD readings were done. The OD was measured on some different concentrations of pure tryptophan solutions and they showed that there were no real big differences in absorbance, even with a 20 time dilution. After some discussion in the group and with supervisors it was unclear how to proceed. Probably it is a bad idea to try to measure the consumptions of tryptophan since it, along with all the proteins in the cell free extract, responds at a wave length of approximately 280 nm. A third enzymatic assay was performed in order to get a free cell extract with a high protein concentration that could be stored. We managed to get one with a good concentration and this is then to be used later on, when we have planned for another way of performing the enzymatic assay. | ||
| + | |||
| + | =='''Week 7, 16th - 20th of July'''== | ||
| + | This has been an eventful week. On Monday we got our sequences back from GATC Biotech and thankfully, the sequences for our cloned genes seem to be correct. Another fun event this week was that we were interviewed by the very nice Bethan Wolfenden from the UCL iGEM team. It was very nice to have the opportunity to talk to another iGEM team and the interview will be broadcasted on the UCL radio show. | ||
| + | |||
| + | '''The receptor group'''<br> | ||
| + | [[File:Lyticase50.png|thumb|200px|Assessment of cell wall degradation.]] | ||
| + | |||
| + | This week, we developed and evaluated a method for degrading the yeast cell wall. Since the cell wall might prevent hCG to reach our receptor, we want to partially remove it before adding the hormone to our cells. For this purpose we chose to use lyticase, which is a mixture of enzymes that degrade the glucans of the yeast cell wall. However, we needed to determine how much lyticase that should be added to the cells and for how long the enzyme should be active to obtain a suitable proportion of protoplasts (cells without cell walls). This was done by adding lyticase to our yeast and then continually measuring OD during the following 60 minutes. Although the experiment had to be repeated once, we now have decided on the proper amount of enzyme and incubation time to use. Another positive result was that the cell wall of the ΔCWP2 strain was degraded faster than the one of the unmodified strain during this assay. This is the first observation that indicates that the cell wall of the ΔCWP2 really is weakened. | ||
| + | <!--The results is presented under [https://2012.igem.org/Team:Chalmers-Gothenburg/Results Results]--> | ||
| + | |||
| + | Our plan was then to add hCG to our cells on Friday but it turned out that our O/N culture of the strains containing yLHCGR, hLHCGR and pSP-GM1 had not grown enough. We decided that the hCG experiment will have to wait until Monday next week. | ||
| + | |||
| + | '''The indigo group''' | ||
| + | |||
| + | This week began with the extraction of the pFig-Indigo from our transformed DH5α from last week. The rest of the week was all about planning and performing the enzymatic assay. Since the strain CEN PK 113-11C with the empty plasmid pIYC04 had now fully grown, this was now used as a control in all experiments. However, the enzymatic assay experiment failed, due to malfunctioning spectrophotometer, and it was decided that we will try it next week again. | ||
| + | Some strains with the pIndigo were also cultivated in a medium with galactose as the main carbon source. This was performed because some literature studies had been done, which implied that glucose may perhaps be an inhibitor for our wanted reaction. | ||
| + | |||
| + | '''The cell wall group'''[[File:Calcofluor_difference.jpg|thumb|300px|Calcofluorstaining of the ''cwp2'' deletion strain compared to the IMFD-73.]] | ||
| + | |||
| + | This week, we performed Calcofluor staining with the cwp2 deletion strain. As a control, the IMFD-73 was used. There was no visible difference in the cell wall between the deletion strain and the control strain. But we got some nice pictures. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =='''Week 6, 9th - 13th of July'''== | ||
| + | [[File:Sekvensering.JPG|thumb|200px|right|Sending off our plasmids for sequencing.]] | ||
| + | Lots of work was done this week! We sent in our plasmids (pIndigo, phLHCGR and pyLHCGR) for sequencing to GATC Biotech. We are keeping our fingers crossed that our work performed in the last 6 weeks has not been in vain and that our sequences are correct. Hopefully we will have the results in the beginning of next week. | ||
| + | |||
| + | '''Receptor group''' | ||
| + | |||
| + | We did not have much to do in the lab this week. Since we now have our IMFD-73 strain transformed with both pYLHCGR and phLHCGR we are actually ready to examine if the cells can detect hCG. Because our IMFD-73 strain has a GFP gene under the control of the FIG1 promoter, we will be able to see if our receptor is functioning by screening for fluorescence after adding hCG. However, this had to wait since we were still waiting for our ordered hCG hormone to arrive and we will also need to develop a strategy for removing the yeast cell wall before adding hCG the first time. Therefore, this week, we mainly focused on developing the wiki page. | ||
'''Indigo group''' | '''Indigo group''' | ||
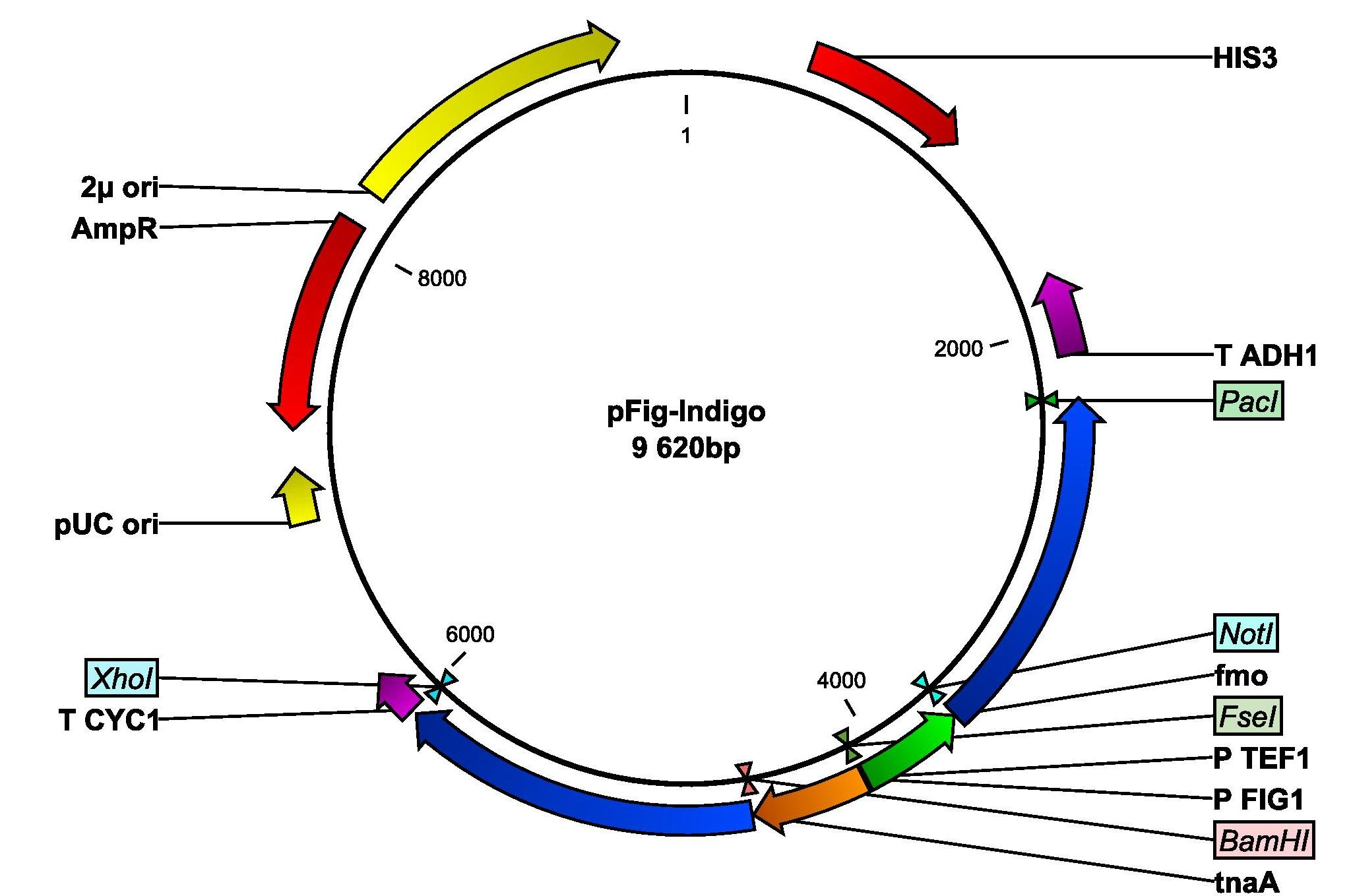
| + | [[File:PFigIndigo.jpg|thumb|200 px|align="right"|The pFigIndigo plasmid.]] | ||
| + | Since no blue color could be observed in our yeast cells that had been transformed with the indigo-synthesizing genes, the indigo group began their week by examining if the lack of indigo production could be due to the low intracellular levels of tryptophan. Since tryptophan is the precursor from which indigo is produced, we tried to grow our cells in different concentrations of the amino acid. Unfortunately, this did not help. We are now discussing how we can proceed with the indigo production. The first thing we are going to try next week is to grow the cells in room temperature. The idea is to give the yeast cells more time to properly fold the proteins. Next week we will also do an enzymatic assay to see if any functioning indigo-synthesizing enzymes are present in the indigo yeast. | ||
| + | Additionally, we also amplified the FIG1 promoter from yeast genomic DNA and we inserted the promoter into our pIndigo plasmid. The new plasmid is now called pFig-Indigo and contains both genes needed for indigo production, whereof one is placed under the control of the FIG1 promotor. DH5α was then transformed with pFig-Indigo. | ||
| + | |||
| + | '''Cell wall group''' | ||
| + | [[File:Cwp2-bp.jpg|thumb|200px|right| The ΔCWP2 strain under the microscope.]] | ||
| + | The cell wall group verified the CWP2 deletion by running colony PCR with our deletion strain. Gel electrophoresis confirmed that the CWP2 gene is gone! Hopefully, we now have a yeast strain with a weakened cell wall that will enable the hCG hormone to pass through it. We also got a close check up on our ΔCWP2 yeast cells under the microscope. No difference in cell shape could be observed when comparing to the normal lab strain. Next week we are going to stain the cell wall and see if it is possible then to identify any differences. | ||
| + | |||
| + | Another work performed this weeke was the transformation of the ΔCWP2 strain with the yLHCGR and hLHCGR plasmids. On Monday we will know if the transformations were successful. | ||
| + | |||
| + | |||
| + | |||
| + | =='''Week 5, 2nd - 6th of July'''== | ||
| + | |||
| + | [[File:Olsson.jpg|right|frame|Helén is preparing the gel electrophoresis.]] | ||
| + | This week the indigo, the receptor and the cell wall group had very similar tasks to perform so we were all reunited in the laboratory. Firstly, on Monday we found out that all transformations of yeast that had been carried out on Friday last week were successful. We now have the IMFD-73 yeast strain transformed with the hLHCGR and yLHCGR plasmid respectively and the cell wall group managed to transform IMFD-73 with both our constructed bipartite fragments and the kanMX fragment. | ||
| + | |||
| + | Unfortunately, even though the cells had been allowed to grow over the weekend, blue cells could still not be seen on the plates that contained yeast transformed with the indigo-synthesizing genes. Hopefully, this is due to the low intracellular levels of tryptophan in the yeast cells, which could prevent proper production of indigo. Therefore, the next step for the indigo group will be to try to add tryptophan to the transformed yeast cells and see if the indigo-production is successful then. But this we will do next week. | ||
| + | |||
| + | [[File:Rachelsofia.jpg|left|frame|Rachel and Sofia are checking the sizes of the fragments.]] | ||
| + | On Monday, we all continued by purifying our transformed yeast cultures by transferring single colonies to new agar plates. On Wednesday the procedure was repeated. On Friday we did a first verification of all our inserts by running colony PCR with the transformed yeast strains. Once again, a big thanks to Microsynth for providing the primers. Gel electrophoresis indicated that inserts are correct in the yLHCGR, hLCGR and the pIndigo plasmid. Results for the kanMX and the bipartite fragments were a bit more disappointing and we are not yet sure if the deletion of the CWP2 gene is successful. | ||
| + | |||
| + | Next week, we will also send in our yLHCGR, hLHCGR and pIndigo plasmid for sequencing and we are really happy to announce that we have a new sponsor, GATC Biotech that has offered to do the sequencing for us. | ||
| + | |||
| + | =='''Week 4, 25th - 29th of June'''== | ||
| + | [[File:Sunshine.jpg|align="right"|frame|Anna, Johan, Rachel and Sofia in the sun.]] | ||
| + | |||
| + | This week, the work of transforming yeast began. Since yeast has a bit slower growth rate than ''E.coli'', we did not always have a lot to do in the lab. But since we were on lab cleaning duty this week, we made sure to keep ourselves busy by running dishwashers, preparing sterile equipment and autoclaving biological waste. We all learned that biowaste, especially in large amounts, does not smell like strawberries. | ||
| + | |||
| + | Amazingly, between some autoclavation program and incubation time we also managed to enjoy a bit of rare sunshine and some icecream. | ||
| + | |||
| + | '''Receptor group''' | ||
| + | |||
| + | We in the receptor group have done three kinds of different tasks this week. The first thing we did was a verification of our plasmids with either the human signal or the yeast signal peptide. We prepared two mixtures with restriction enzymes and a 3 % agarose gel and the gel electrophoresis showed that the inserts were of the correct lengths. Happy! The plasmids were now ready to be used to transform yeast and we continued our work with preparing agar plates that are going to be used for selection. We made -his, -ura and -his-ura agar plates. | ||
| + | |||
| + | In the second half of the week we transformed our special yeast strain IMFD-73 (optimized for the expression of G protein-coupled receptors), provided by the Kondo group from Kobe University, with our plasmids hLHCGR, yLHCGR and the empty expression plasmid pSP-GM1. On Friday we plated the yeast on -ura plates and next week on Monday we will see if the transformation has worked. | ||
| + | [[File:hLHCGR2.jpg|center|frame|Our hLHCGR plasmid containing the LHCGR gene with the human signal peptide.]] | ||
| + | |||
| + | |||
| + | |||
| + | '''Indigo group''' | ||
| + | |||
| + | This week was pretty eventless and the Indigo group spent a lot of time waiting for incubations of cells. We purified the plasmid pIndigo from the successful transformation from week 3. During wednesday, the yeast strain CEN.PK 113-11C was transformed with the plasmid and the cells were plated onto SD His- plates. We had hoped that there would be blue colonies, i.e. indigo synthesizing cells, on the plates on Friday but only regular colonies could be seen. The Indigo group is now hoping that the cells will turn blue during the weekend, otherwise we will try to add the precursor of the indigo reaction, tryptophan. The pIndigo will also be sent for sequencing during week 5 to ensure that the inserts are right. | ||
| + | |||
| + | [[File:Gelelectrophoresis.jpg|align="right"|width="60"|frame|Rachel is loading one of many gels.]] | ||
| + | |||
| + | |||
| + | '''Cell wall group''' | ||
| + | |||
| + | This week, the yeast strains for our back-up plan arrived. This strain lacks the ''CWP2'' gene and has the kanamycin resistance cassette inserted instead. As a back-up plan, in case the Bipartite method will not work, we can amplify the kanMX cassette from this strain together with the 500 bp up- and downstream region of the ''CWP2'' gene and then transform our yeast strain with this fragment. The advantage with this method is that we only have to insert one fragment instead of two into our yeast strain. So, colony PCR was performed with the Δ''cwp2'' strain and in contrary to expectations, it worked directly and we got a nice band of the right size on th gel. The fragment was then purified from the gel. | ||
| + | |||
| + | In the second half of the week, our special yeast strain IMFD-73 was transformed with the whole kanMX fragment. Another transformation was done with the two Bipartite fragments. The cells were then plated on G418 plates. Next week we will see if the transformation was successful in one or both of the cases. | ||
| + | |||
| + | =='''Week 3, 18th - 22th of June'''== | ||
| + | |||
| + | This week Chalmers' iGEM team really worked hard. Early mornings and late nights. Our hands almost bled from all the pipetting... Okay, not really. But we actually made some great progress this week. Check out details below![[File:Ge_small.jpg|align="right"|width="60"|frame|You don't always get the results you expect from gel electrophoresis... We tried to verify that our yLHCGR construct is correct but did not quite know how to interpret our generated results. However, after a second round we could conclude that the construct is correct. ]] | ||
| + | |||
| + | |||
| + | '''Receptor group''' | ||
| + | |||
| + | We began our week with transforming ''E. coli'' DH5α with our LHCGR constructs (that had either a yeast or human signal peptide) that were ligated into the pSP-GM1 plasmid. ''E. coli'' was then plated onto LB Amp agar plates and on Tuesday we were very happy to see that at least cells transformed with the human version of the LHCGR were happily growing. We proceeded by purifying the plasmids, which will now be called hLHCGR, from these cells. | ||
| + | |||
| + | To make certain that our constructed LHCGR fragment with the yeast signal (that we tried to insert into the pSP-GM1 plasmid) is correct, we used gel electrophoresis. From this we concluded that our yLHCGR fragment is correct and therefore we attempted to ligate this fragment into the pSP-GM1 once again. We transformed DH5α with this ligated plasmid and the next day, to our great happiness, we discovered colonies of transformed DH5α growing on the LB amp agar plates. Hopefully (verification still need to be done), we now have both versions of the LHCGR gene ligated into pSP-GM1, just waiting to be transformed into yeast . Great success! We finished our week by purifying the new plasmid, which hereafter will be called yLHCGR, from the DH5α cells. | ||
| + | |||
| + | |||
| + | '''Indigo group''' | ||
| + | [[File:week3S.jpg|left|frame|Sofia made the ligation mixtures for the Indigo group.]] | ||
| + | |||
| + | On Monday two transformations were made from the ligation mixtures that have been prepared in week 2. On Tuesday we could see that our transformations had been successful and colonies were picked from the plates to inoculate. The pIYC04+fmo plasmid and the pIYC04+tnaA plasmid were purified from the cultures, digested and then purified on gel. The pIYC04+fmo plasmid was ligated with the tnaA gene and the pIYC04+tnaA with the fmo gene. Two different transformations were made from the ligation mixtures and on Friday we could see that one of them was successful. So now we have cells containing the plasmid, pIndigo, with both the fmo and the tnaA gene and that is supposed to be inserted into yeast to enable the bio-indigo production. | ||
| + | |||
| + | |||
| - | |||
| - | We made a ligation mixture of the fmo gene with pIYC04 and tried to transform the ligation mix into E. coli. Unfortunately nothing had grown on the ligation plate (nor the control plate) and we had to redo the ligation. So the last day of the week we did two ligations in parallel, one with the fmo gene and pIYC04 and another one with the tnaA gene and pIYC04. These will be transformed into E. coli next week and hopefully one of them will work then. | + | |
| + | |||
| + | '''The cell wall group''' | ||
| + | |||
| + | The highlight of the week was the article in the newspaper Metro about our project and about the head of the cell wall group, Rachel. The article can be found [http://allastudier.se/artiklar/865-labbet-ist%C3%A4llet-f%C3%B6r-stranden-i-sommar/ here]. | ||
| + | Besides answering question of a journalist, the goal for this week was to fuse together the upstream fragment of the CWP2 gene with the 5’ end of the kanamycin resistance cassette and the downstream fragment of the CWP2 gene with the 3’ end of the kanamycin resistance cassette. To fuse together the fragments turned up to be even more tricky than expected. Several PCR reactions were run this week under different conditions such as different temperatures, different extending times and gradually increasing temperatures. Another attempt was to first only run 10 cycles with only the overlapping fragments and adding the primers afterwards. But all these trials resulted in the same outcome: One clear band at 1000 bp for the first fusion product and a smear for the second fusion product. But the expected sizes for both fragments were 1500 bp. This lead to slightly frustrated groupmembers (Citat Rahul: "How pissed off is Rachel really?"). But of course, we did not give up! What has finally saved us was the advice of PCR Pro Il-Kwon, the nice Korean guy in the System Biology group that already helped us before to get the fmo gene. He suggested another temperature, the use of a little less enzyme and the checking of the PCR-products on a GelRed gel first instead purifying it directly on a GelGreen gel. And it worked! GelRed gives much more precise results when checking the length of the fragments and it is not quite unlikely that we got right results already in the previous approaches but that we could not see them properly on the GelGreen gel. Nevertheless, we were very happy to finally succeed with the Fusion PCR and to get the two fragments that we will transform yeast with next week. Stay tuned! | ||
| + | |||
| + | [[File:Bipartite2.jpg|center|frame|For those of you that do not have all our methods in mind. This is what we want to do in order to delete the cell wall protein gene. The PCR1 reactions we did last week, this week we want to fuse together the fragments.]] | ||
| + | |||
| + | =='''Week 2, 11th - 15th of June''' == | ||
| + | |||
| + | This week, the real lab work began. In order to create the biodegradable pregnancy test kit, we have three main tasks to solve. These are 1) the expression of the receptor, 2) the insertion of the indigo producing enzymes and 3) the deletion of a cell wall protein in order to enhance the chances of the ligand to pass the cell wall. Therefore we divided ourselves into three groups, the “receptor group”, the “cell wall group” and the “indigo group” each responsible for its task. Running PCR was a big part of the work this week and we would like to thank Microsynth AG for sponsoring us with the primers. | ||
| + | |||
| + | |||
| + | '''Receptor group''' | ||
| + | |||
| + | This week, we ran PCR with our lhcgr primers from Microsynth together with our LHCGR construct. This was done in order to obtain the gene for the LH/CG receptor fused with the yeast signal peptide of the LHCGR construct. We religated our generated PCR product and then transformated ''E. coli'' DH5α with it. We also did a control where E. coli was transformed with the non-ligated PCR product. We were very pleased to find that our cells that were transformed with the ligated LHCGR construct were growing very nicely on the LB-Amp plate while the control cells did not. | ||
| + | |||
| + | We purified the LHCGR construct from the transformed ''E. coli'' and we then continued our work with cutting out the LHCGR gene, that should now be fused with the yeast signal, from the religated construct and [[File:PlatesLHCGR.jpg|left|frame|Cells are dead on the control plate but colonies are growing on the LHCGR plate. Nice!]] ligate it into our shuttle plasmid pSP-GM1. Simultaneously, the original LHCGR construct was also cut to generate the LHCGR gene with its associated endogenous human signal peptide. After cutting we ran gel electrophoresis to verify correct fragment sizes and the DNA from the bands containing the LHCGR gene with the human respectively yeast signal peptide were extracted and purified along with the cut pSP-GM1. Finally, we also attempted to ligate the two LHCGR inserts into the pSP-GM1 plasmids. Will the ligation work? This we will need to find out next week! | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''Indigo group''' | ||
| + | [[File:JohanBiö.jpg |align="right"|frame|Johan is analyzing the results.]] PCR reactions were run for the tnaA and the fmo gene. The tnaA gene was taken directly from ''E. coli'' with colony-PCR whereas the fmo gene was kindly provided by Si Wouk Kim from Chosun University, South Korea. After two days we succeeded to purify the fmo gene from gel. When the tnaA gene did not show up on the gel after the two first days we got the advice from Rahul Kumar (who works here in the lab at the department of System’s biology at Chalmers University of Technology) to boil the ''E. coli'' cells in a basic solution before the colony PCR. This time the colony PCR worked and the tnaA gene could be purified from the gel. Thanks Rahul! | ||
| + | |||
| + | We made a ligation mixture of the fmo gene with pIYC04 and tried to transform the ligation mix into ''E. coli''. Unfortunately nothing had grown on the ligation plate (nor the control plate) and we had to redo the ligation. So the last day of the week we did two ligations in parallel, one with the fmo gene and pIYC04 and another one with the tnaA gene and pIYC04. These will be transformed into ''E. coli'' next week and hopefully one of them will work then. | ||
| Line 46: | Line 273: | ||
We started with the construction of the knock-out fragments. First, we ran PCR with genomic yeast DNA in order to get the 500 up- and downstream fragments of the CWP2 gene. The two overlapping kanMX fragments were also amplified by PCR. For these PCR reactions, we used HPLC purified primers that were sponsored by Microsynth AG. After PCR the fragments were checked and purified on gel. | We started with the construction of the knock-out fragments. First, we ran PCR with genomic yeast DNA in order to get the 500 up- and downstream fragments of the CWP2 gene. The two overlapping kanMX fragments were also amplified by PCR. For these PCR reactions, we used HPLC purified primers that were sponsored by Microsynth AG. After PCR the fragments were checked and purified on gel. | ||
| - | We have learned that everything takes a little bit longer than expected; it took actually the whole week to get all the four fragments, since some PCR reaction had to be repeated several times. However, we are still on schedule. On Friday, we could then start with Fusion PCR in order to fuse the fragments. Of course it did not work the first time and we have the feeling that this will be the more tricky part of the cell wall deletion. But we will come up with some different approaches next week! | + | We have learned that everything takes a little bit longer than expected; it took actually the whole week to get all the four fragments, [[File:PCR2.jpg|left|frame|The moment of truth: Did the PCR work this time?]]since some PCR reaction had to be repeated several times. However, we are still on schedule. On Friday, we could then start with Fusion PCR in order to fuse the fragments. Of course it did not work the first time and we have the feeling that this will be the more tricky part of the cell wall deletion. But we will come up with some different approaches next week! |
| + | |||
| + | |||
| + | |||
| + | |||
| + | =='''Week 1, 4th - 8th of June'''== | ||
| + | |||
| + | '''Monday, 4th of June''' | ||
| + | [[File:Rachel55.jpg|align="right"|frame|Rachel is especially happy to get on with the laboratory work.]][[File:Anna2.jpg|align="right"|frame|Anna working with the purification of the plasmids.]]The laboratory work has begun and after preparing this work for several months we are all very excited to finally get it started. Today we started preparing sterile equipment and we also wrote risk declarations for all of our planned experiments. We also introduced ourselves and held a brief presentation of our project to the very nice people working at the department of System’s biology here at Chalmers University of Technology. | ||
| + | |||
| + | '''Tuesday, 5th of June''' | ||
| + | |||
| + | Today our dear yeast strains that were kindly provided to us by the Kondo research group of Kobe University, Japan, were transferred to new and fresh YPD agar plates. We also prepared buffers needed for making competent'' E. coli'' and we inoculated ''E. coli'' DH5α in LB medium. | ||
| + | |||
| + | '''Wednesday, 6th of June''' | ||
| + | |||
| + | We prepared a stock of competent ''E. coli'' DH5α. Today we also amplified the shuttle plasmids pSP-GM1 and pIYC04 that we will use in our laboratory work, by transforming ''E. coli'' DH5α. The cells were then grown on LB-Amp agar plates. We also worked with the team wiki page and created a banner and a logo for the site. | ||
| + | |||
| + | '''Thursday, 7th of June''' | ||
| + | |||
| + | Not much laboratory could be done today. Two colonies from each plate containing the ''E. coli'' DH5α that were transformed with either pSP-GM1 or pIYC04, were inoculated in ampicillin-containing media and were grown overnight. For the rest of the day, we worked with creating a team brochure and we also established a Facebook page for the team. Click [http://www.facebook.com/ChalmersGothenburgiGEMTeam2012 here] to visit our page and Like our team! | ||
| + | |||
| + | '''Friday, 8th of June''' | ||
| + | |||
| + | We purified our pSP-GM1 and pIYC04 plasmids from the transformed ''E. coli''. | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
{{Team:Chalmers-Gothenburg/footer}} | {{Team:Chalmers-Gothenburg/footer}} | ||
Latest revision as of 20:17, 26 September 2012
Week 13 - Week 17, 27th of August - 26th of September
During these weeks, we were very busy with paper work, such as the summary, the analysis and discussion of our results and the update of the wiki page. We also started to prepare the presentation of our project for the Jamboree.
In addition, we continued with the cloning of our parts into BioBricks and we managed finally to send in the FIG1 promoter. Another task was the preparation of glycerol stocks of our yeast strains and lastly we had to clean the lab.
SEE YOU IN AMSTERDAM!
Week 12, 20th - 24th of August
BioBrick construction
We spent most of the time on Miniprep and gelelectrophoresis in order to prepare and verify our BioBrick vectors containing our parts. But the results on the gels were not satisfying so we redid the ligation and transformation once more. Next week we hopefully can finish our BioBricks and send it to the Registry.
Indigo assay
We decided to do one last experiment with our strains containing the indigo genes. The goal of this experiment was to investigate which compounds that are present in these colourful strains. The strain was grown in shake flasks in both galactose and glucose and after 24h, 2g/l of tryptophan was added. After another 24h, the same redbrown respectively greenbrown colonies as in week 10 were observed. We centrifuged the cells and measured the absorbance spectrum of the colorful supernatant. Some peaks could be observed and next week, we will analyze the spectrum in more detail och hopefully be able to explain the arising colours.
The rest of the time we used for paper work such as summarizing our results, preparing the presentation for the Jamboree and designing the poster.
Week 11, 13th - 17th of August
Final hCG test
One final hCG test was performed. This time the whole system i.e. the yeast strain with the deleted CWP2 gene and containing both the LHCG-receptor and the indigo genes (with the monooxygenase expressed constitutively and the tryptophanase coupled with the pheromone pathway through the FIG1 promoter) was tested. The strain was grown in SD –His –Ura medium with additional tryptophan (2g/l) and hCG hormone was added.
If the system would work, the hormone should activate the pheromone pathway which in turn should activate the expression of tryptophanase. This would then result in digestion of tryptophan and the same brown colour, which has been observed in the strains containing the activated indigo genes, should appear. Unfortunately, no colour change could be observed. That means that the whole system is not working properly. This could be due the limited permeability of the cell wall or a malfunctioning receptor.
BioBricks
Besides testing our yeast strain, the task of constructing our BioBricks vectors was time-consuming this week. After another unsuccessful transformation, we discovered that we had used the wrong selection antibiotic. Haha, no wonder that no single colony could be observed! Well, we prepared new plates, this time with chloramphenicol, and repeated the digestion/ligation/transformation. And finally, we got some colonies! Next week, we will purify the plasmids and hopefully send it to the Registry of Standard Biological Parts.
Human practices
A big event of this week was the presentation about Biotechnology and our project for secondary school and high school students. They were participating in a summer research course. It was a real challenge to explain what we are doing for students that barely know what DNA is. But it was fun and we think the students really liked our presentation.
Week 10, 6th - 10th of August
Receptor group
This week we did a last hCG test in which we screened for fluorescence in the cells that we added the hCG hormone to. This time three other strains were used: IMFD-70, IMFD-72 and IMFD-74 and they had all been transformed with LH/CG receptor. These strains contain different types of Gα proteins and our hopes were that these would enable the yeast cells to better couple the LH/CG receptor with the pheromone pathway. However, when doing the hCG testing procedure no significant difference could be observed when examining the cells in the fluorescence microscope: all cells, no matter if they had had the hormone added to them or not, were displaying a weak fluorescence. As before, we are not sure whether or not our receptor is functioning and actually detecting the hormone or if the fluorescence displayed by the cells that have been treated with hCG is only background noise.
The last thing that we are going to do now is to add hormone to our completed strain that expresses both the LH/CG receptor and the indigo genes. This is our last shot at testing to see if the receptor is functioning. We will do the test next week and we would really like to see some of that color-change that we previously have observed in the indigo strain but now in response to detecting the hormone!
Construction of BioBricks
We decided to register three new parts, the LHCGR gene with two different signal peptides and the FIG1 promoter. This week, after receiving the necessary primers from Microsynth AG, we started with the cloning of our parts into the BioBrick vector. The first step was PCR amplification of the three parts. We have become experienced in PCR conditions during this summer and we succeeded directly with the PCR in the first approach. The fragments were purified from gel and digested with PstI and EcoRI for 30 minutes. The vector was also digested with the same enzymes and DpnI additionally, as described in the protocol for BioBricks construction provided by Registry of Standard Biological Parts. The reaction was terminated by putting the samples on 80° C for 10 minutes. A ligation mix with insert, vector, T4 ligase and ligation buffer was prepared for each of the three parts. As a negative control, water instead of insert was added to the mix. The samples were incubated for 3h in room temperature and after that, E.coli was transformed with the hopefully correctly ligated constructs. The anticlimax arose when we checked the plates the day after. Nothing has grown! The only positive thing was that nothing has grown on the negative control either. We tried to investigate what could have gone wrong. We know that ligation uses to be tricky but we have been very successful earlier and it was actually the first time we did not succeed with ligation. This time we followed the protocol for the BioBrick construction and this means that we did not purify the DNA after digestion. Instead, we terminated the digestion by heating the samples and then proceeded directly with ligation. Another big difference was the incubation time for ligation. In our previous approaches, we have always ligated over night, this time, we only incubated the ligation mix for 3 hours. So next week, we will just use our own protocol again. Let’s hope it will work!
Week 9, 30th of July - 3rd of August
The complete system has now been put together! The IMFD-73 strain that has had the CWP2 gene deleted is now expressing the LH/CG receptor and has been transformed with both genes needed for indigo production, one of them being placed under the control of the pheromone-induced FIG1 promoter. Will this system be able to function as a pregnancy test? This is a question that we still need to answer. This week, we therefore continued to test our receptor by adding hormone to our yeast cells.
Receptor group
In contrast with last week, we grew our cells in room temperature before adding hCG. This time we also used the ΔCWP2 strain, which we didn’t treat with any cell wall degrading enzymes before the addition of the hormone. Sadly, approximately the same observations as last week could be made. When examining our cells in the fluorescence microscope, all cells displayed a weak fluorescence, both cells that had been treated with the hormone and cells that had not.
In parallel to the hCG tests that have been carried out, we have now also transformed three other strains with the LH/CG receptor. Next week, these strains will be used in our hCG assay instead and we really hope that these will show some more promising results.
Week 8, 23rd - 27th of July
This week, we focused on investigating our yeast strains that had been transformed with different plasmids. Different assays were performed as described below.
Receptor group
This week was very exciting but also a bit nerve-racking for us in the receptor group since this was the week when we actually got to test our receptor. On Tuesday, we degraded the cell wall of the IMFD-73 strain that had been transformed with the yLHCGR plasmid and the hLHCGR plasmid. As a control, the strain transformed with the empty pSP-GM1 plasmid was also treated. The hCG hormone was then added to the cells.
The IMFD-73 strain that we have used to express our receptor in, has a GFP gene placed under the control of the FIG1 promoter. So, if the receptor is functioning, the hCG hormone should be able to activate the yeast pheromone pathway and thus induce the expression of the GFP gene. Therefore, we used a fluorescence microscope to examine our cells. To our great disappointment, the level of fluorescence was approximately the same when comparing cells that had been treated with the hormone and cells that had not been treated with the hormone. We were already aware of the risk of having levels of background signaling in our IMFD-73 strain and since the fluorescence intensity was approximately the same in the hormone-treated and the non-treated cells, chances are that our receptor is not functioning and is not producing any output signal.
There is still hope though! Next week we will try to grow our yeast in room temperature to hopefully give the cells better chances to express and properly fold the receptor. We will also try to express our receptor in three other strains that also have been optimized for expression of mammalian GPCRs but that are a bit different from the IMFD-73 strain that we have used so far.
Indigo group
This week was all about the enzymatic assay on tryptophanase. In total there were three of these performed this week and the results were not that satisfying. The readings of the first one did not show any reliable results on consumption of tryptophan. The second one ended up with a too low concentration of proteins in the cell free extract, so no OD readings were done. The OD was measured on some different concentrations of pure tryptophan solutions and they showed that there were no real big differences in absorbance, even with a 20 time dilution. After some discussion in the group and with supervisors it was unclear how to proceed. Probably it is a bad idea to try to measure the consumptions of tryptophan since it, along with all the proteins in the cell free extract, responds at a wave length of approximately 280 nm. A third enzymatic assay was performed in order to get a free cell extract with a high protein concentration that could be stored. We managed to get one with a good concentration and this is then to be used later on, when we have planned for another way of performing the enzymatic assay.
Week 7, 16th - 20th of July
This has been an eventful week. On Monday we got our sequences back from GATC Biotech and thankfully, the sequences for our cloned genes seem to be correct. Another fun event this week was that we were interviewed by the very nice Bethan Wolfenden from the UCL iGEM team. It was very nice to have the opportunity to talk to another iGEM team and the interview will be broadcasted on the UCL radio show.
The receptor group
This week, we developed and evaluated a method for degrading the yeast cell wall. Since the cell wall might prevent hCG to reach our receptor, we want to partially remove it before adding the hormone to our cells. For this purpose we chose to use lyticase, which is a mixture of enzymes that degrade the glucans of the yeast cell wall. However, we needed to determine how much lyticase that should be added to the cells and for how long the enzyme should be active to obtain a suitable proportion of protoplasts (cells without cell walls). This was done by adding lyticase to our yeast and then continually measuring OD during the following 60 minutes. Although the experiment had to be repeated once, we now have decided on the proper amount of enzyme and incubation time to use. Another positive result was that the cell wall of the ΔCWP2 strain was degraded faster than the one of the unmodified strain during this assay. This is the first observation that indicates that the cell wall of the ΔCWP2 really is weakened.
Our plan was then to add hCG to our cells on Friday but it turned out that our O/N culture of the strains containing yLHCGR, hLHCGR and pSP-GM1 had not grown enough. We decided that the hCG experiment will have to wait until Monday next week.
The indigo group
This week began with the extraction of the pFig-Indigo from our transformed DH5α from last week. The rest of the week was all about planning and performing the enzymatic assay. Since the strain CEN PK 113-11C with the empty plasmid pIYC04 had now fully grown, this was now used as a control in all experiments. However, the enzymatic assay experiment failed, due to malfunctioning spectrophotometer, and it was decided that we will try it next week again. Some strains with the pIndigo were also cultivated in a medium with galactose as the main carbon source. This was performed because some literature studies had been done, which implied that glucose may perhaps be an inhibitor for our wanted reaction.
The cell wall groupThis week, we performed Calcofluor staining with the cwp2 deletion strain. As a control, the IMFD-73 was used. There was no visible difference in the cell wall between the deletion strain and the control strain. But we got some nice pictures.
Week 6, 9th - 13th of July
Lots of work was done this week! We sent in our plasmids (pIndigo, phLHCGR and pyLHCGR) for sequencing to GATC Biotech. We are keeping our fingers crossed that our work performed in the last 6 weeks has not been in vain and that our sequences are correct. Hopefully we will have the results in the beginning of next week.
Receptor group
We did not have much to do in the lab this week. Since we now have our IMFD-73 strain transformed with both pYLHCGR and phLHCGR we are actually ready to examine if the cells can detect hCG. Because our IMFD-73 strain has a GFP gene under the control of the FIG1 promoter, we will be able to see if our receptor is functioning by screening for fluorescence after adding hCG. However, this had to wait since we were still waiting for our ordered hCG hormone to arrive and we will also need to develop a strategy for removing the yeast cell wall before adding hCG the first time. Therefore, this week, we mainly focused on developing the wiki page.
Indigo group
Since no blue color could be observed in our yeast cells that had been transformed with the indigo-synthesizing genes, the indigo group began their week by examining if the lack of indigo production could be due to the low intracellular levels of tryptophan. Since tryptophan is the precursor from which indigo is produced, we tried to grow our cells in different concentrations of the amino acid. Unfortunately, this did not help. We are now discussing how we can proceed with the indigo production. The first thing we are going to try next week is to grow the cells in room temperature. The idea is to give the yeast cells more time to properly fold the proteins. Next week we will also do an enzymatic assay to see if any functioning indigo-synthesizing enzymes are present in the indigo yeast. Additionally, we also amplified the FIG1 promoter from yeast genomic DNA and we inserted the promoter into our pIndigo plasmid. The new plasmid is now called pFig-Indigo and contains both genes needed for indigo production, whereof one is placed under the control of the FIG1 promotor. DH5α was then transformed with pFig-Indigo.
Cell wall group
The cell wall group verified the CWP2 deletion by running colony PCR with our deletion strain. Gel electrophoresis confirmed that the CWP2 gene is gone! Hopefully, we now have a yeast strain with a weakened cell wall that will enable the hCG hormone to pass through it. We also got a close check up on our ΔCWP2 yeast cells under the microscope. No difference in cell shape could be observed when comparing to the normal lab strain. Next week we are going to stain the cell wall and see if it is possible then to identify any differences.
Another work performed this weeke was the transformation of the ΔCWP2 strain with the yLHCGR and hLHCGR plasmids. On Monday we will know if the transformations were successful.
Week 5, 2nd - 6th of July
This week the indigo, the receptor and the cell wall group had very similar tasks to perform so we were all reunited in the laboratory. Firstly, on Monday we found out that all transformations of yeast that had been carried out on Friday last week were successful. We now have the IMFD-73 yeast strain transformed with the hLHCGR and yLHCGR plasmid respectively and the cell wall group managed to transform IMFD-73 with both our constructed bipartite fragments and the kanMX fragment.
Unfortunately, even though the cells had been allowed to grow over the weekend, blue cells could still not be seen on the plates that contained yeast transformed with the indigo-synthesizing genes. Hopefully, this is due to the low intracellular levels of tryptophan in the yeast cells, which could prevent proper production of indigo. Therefore, the next step for the indigo group will be to try to add tryptophan to the transformed yeast cells and see if the indigo-production is successful then. But this we will do next week.
On Monday, we all continued by purifying our transformed yeast cultures by transferring single colonies to new agar plates. On Wednesday the procedure was repeated. On Friday we did a first verification of all our inserts by running colony PCR with the transformed yeast strains. Once again, a big thanks to Microsynth for providing the primers. Gel electrophoresis indicated that inserts are correct in the yLHCGR, hLCGR and the pIndigo plasmid. Results for the kanMX and the bipartite fragments were a bit more disappointing and we are not yet sure if the deletion of the CWP2 gene is successful.
Next week, we will also send in our yLHCGR, hLHCGR and pIndigo plasmid for sequencing and we are really happy to announce that we have a new sponsor, GATC Biotech that has offered to do the sequencing for us.
Week 4, 25th - 29th of June
This week, the work of transforming yeast began. Since yeast has a bit slower growth rate than E.coli, we did not always have a lot to do in the lab. But since we were on lab cleaning duty this week, we made sure to keep ourselves busy by running dishwashers, preparing sterile equipment and autoclaving biological waste. We all learned that biowaste, especially in large amounts, does not smell like strawberries.
Amazingly, between some autoclavation program and incubation time we also managed to enjoy a bit of rare sunshine and some icecream.
Receptor group
We in the receptor group have done three kinds of different tasks this week. The first thing we did was a verification of our plasmids with either the human signal or the yeast signal peptide. We prepared two mixtures with restriction enzymes and a 3 % agarose gel and the gel electrophoresis showed that the inserts were of the correct lengths. Happy! The plasmids were now ready to be used to transform yeast and we continued our work with preparing agar plates that are going to be used for selection. We made -his, -ura and -his-ura agar plates.
In the second half of the week we transformed our special yeast strain IMFD-73 (optimized for the expression of G protein-coupled receptors), provided by the Kondo group from Kobe University, with our plasmids hLHCGR, yLHCGR and the empty expression plasmid pSP-GM1. On Friday we plated the yeast on -ura plates and next week on Monday we will see if the transformation has worked.
Indigo group
This week was pretty eventless and the Indigo group spent a lot of time waiting for incubations of cells. We purified the plasmid pIndigo from the successful transformation from week 3. During wednesday, the yeast strain CEN.PK 113-11C was transformed with the plasmid and the cells were plated onto SD His- plates. We had hoped that there would be blue colonies, i.e. indigo synthesizing cells, on the plates on Friday but only regular colonies could be seen. The Indigo group is now hoping that the cells will turn blue during the weekend, otherwise we will try to add the precursor of the indigo reaction, tryptophan. The pIndigo will also be sent for sequencing during week 5 to ensure that the inserts are right.
Cell wall group
This week, the yeast strains for our back-up plan arrived. This strain lacks the CWP2 gene and has the kanamycin resistance cassette inserted instead. As a back-up plan, in case the Bipartite method will not work, we can amplify the kanMX cassette from this strain together with the 500 bp up- and downstream region of the CWP2 gene and then transform our yeast strain with this fragment. The advantage with this method is that we only have to insert one fragment instead of two into our yeast strain. So, colony PCR was performed with the Δcwp2 strain and in contrary to expectations, it worked directly and we got a nice band of the right size on th gel. The fragment was then purified from the gel.
In the second half of the week, our special yeast strain IMFD-73 was transformed with the whole kanMX fragment. Another transformation was done with the two Bipartite fragments. The cells were then plated on G418 plates. Next week we will see if the transformation was successful in one or both of the cases.
Week 3, 18th - 22th of June
This week Chalmers' iGEM team really worked hard. Early mornings and late nights. Our hands almost bled from all the pipetting... Okay, not really. But we actually made some great progress this week. Check out details below!
Receptor group
We began our week with transforming E. coli DH5α with our LHCGR constructs (that had either a yeast or human signal peptide) that were ligated into the pSP-GM1 plasmid. E. coli was then plated onto LB Amp agar plates and on Tuesday we were very happy to see that at least cells transformed with the human version of the LHCGR were happily growing. We proceeded by purifying the plasmids, which will now be called hLHCGR, from these cells.
To make certain that our constructed LHCGR fragment with the yeast signal (that we tried to insert into the pSP-GM1 plasmid) is correct, we used gel electrophoresis. From this we concluded that our yLHCGR fragment is correct and therefore we attempted to ligate this fragment into the pSP-GM1 once again. We transformed DH5α with this ligated plasmid and the next day, to our great happiness, we discovered colonies of transformed DH5α growing on the LB amp agar plates. Hopefully (verification still need to be done), we now have both versions of the LHCGR gene ligated into pSP-GM1, just waiting to be transformed into yeast . Great success! We finished our week by purifying the new plasmid, which hereafter will be called yLHCGR, from the DH5α cells.
Indigo group
On Monday two transformations were made from the ligation mixtures that have been prepared in week 2. On Tuesday we could see that our transformations had been successful and colonies were picked from the plates to inoculate. The pIYC04+fmo plasmid and the pIYC04+tnaA plasmid were purified from the cultures, digested and then purified on gel. The pIYC04+fmo plasmid was ligated with the tnaA gene and the pIYC04+tnaA with the fmo gene. Two different transformations were made from the ligation mixtures and on Friday we could see that one of them was successful. So now we have cells containing the plasmid, pIndigo, with both the fmo and the tnaA gene and that is supposed to be inserted into yeast to enable the bio-indigo production.
The cell wall group
The highlight of the week was the article in the newspaper Metro about our project and about the head of the cell wall group, Rachel. The article can be found [http://allastudier.se/artiklar/865-labbet-ist%C3%A4llet-f%C3%B6r-stranden-i-sommar/ here]. Besides answering question of a journalist, the goal for this week was to fuse together the upstream fragment of the CWP2 gene with the 5’ end of the kanamycin resistance cassette and the downstream fragment of the CWP2 gene with the 3’ end of the kanamycin resistance cassette. To fuse together the fragments turned up to be even more tricky than expected. Several PCR reactions were run this week under different conditions such as different temperatures, different extending times and gradually increasing temperatures. Another attempt was to first only run 10 cycles with only the overlapping fragments and adding the primers afterwards. But all these trials resulted in the same outcome: One clear band at 1000 bp for the first fusion product and a smear for the second fusion product. But the expected sizes for both fragments were 1500 bp. This lead to slightly frustrated groupmembers (Citat Rahul: "How pissed off is Rachel really?"). But of course, we did not give up! What has finally saved us was the advice of PCR Pro Il-Kwon, the nice Korean guy in the System Biology group that already helped us before to get the fmo gene. He suggested another temperature, the use of a little less enzyme and the checking of the PCR-products on a GelRed gel first instead purifying it directly on a GelGreen gel. And it worked! GelRed gives much more precise results when checking the length of the fragments and it is not quite unlikely that we got right results already in the previous approaches but that we could not see them properly on the GelGreen gel. Nevertheless, we were very happy to finally succeed with the Fusion PCR and to get the two fragments that we will transform yeast with next week. Stay tuned!
Week 2, 11th - 15th of June
This week, the real lab work began. In order to create the biodegradable pregnancy test kit, we have three main tasks to solve. These are 1) the expression of the receptor, 2) the insertion of the indigo producing enzymes and 3) the deletion of a cell wall protein in order to enhance the chances of the ligand to pass the cell wall. Therefore we divided ourselves into three groups, the “receptor group”, the “cell wall group” and the “indigo group” each responsible for its task. Running PCR was a big part of the work this week and we would like to thank Microsynth AG for sponsoring us with the primers.
Receptor group
This week, we ran PCR with our lhcgr primers from Microsynth together with our LHCGR construct. This was done in order to obtain the gene for the LH/CG receptor fused with the yeast signal peptide of the LHCGR construct. We religated our generated PCR product and then transformated E. coli DH5α with it. We also did a control where E. coli was transformed with the non-ligated PCR product. We were very pleased to find that our cells that were transformed with the ligated LHCGR construct were growing very nicely on the LB-Amp plate while the control cells did not.
We purified the LHCGR construct from the transformed E. coli and we then continued our work with cutting out the LHCGR gene, that should now be fused with the yeast signal, from the religated construct and ligate it into our shuttle plasmid pSP-GM1. Simultaneously, the original LHCGR construct was also cut to generate the LHCGR gene with its associated endogenous human signal peptide. After cutting we ran gel electrophoresis to verify correct fragment sizes and the DNA from the bands containing the LHCGR gene with the human respectively yeast signal peptide were extracted and purified along with the cut pSP-GM1. Finally, we also attempted to ligate the two LHCGR inserts into the pSP-GM1 plasmids. Will the ligation work? This we will need to find out next week!
Indigo group
We made a ligation mixture of the fmo gene with pIYC04 and tried to transform the ligation mix into E. coli. Unfortunately nothing had grown on the ligation plate (nor the control plate) and we had to redo the ligation. So the last day of the week we did two ligations in parallel, one with the fmo gene and pIYC04 and another one with the tnaA gene and pIYC04. These will be transformed into E. coli next week and hopefully one of them will work then.
The cell wall group
We started with the construction of the knock-out fragments. First, we ran PCR with genomic yeast DNA in order to get the 500 up- and downstream fragments of the CWP2 gene. The two overlapping kanMX fragments were also amplified by PCR. For these PCR reactions, we used HPLC purified primers that were sponsored by Microsynth AG. After PCR the fragments were checked and purified on gel.
We have learned that everything takes a little bit longer than expected; it took actually the whole week to get all the four fragments, since some PCR reaction had to be repeated several times. However, we are still on schedule. On Friday, we could then start with Fusion PCR in order to fuse the fragments. Of course it did not work the first time and we have the feeling that this will be the more tricky part of the cell wall deletion. But we will come up with some different approaches next week!
Week 1, 4th - 8th of June
Monday, 4th of June
The laboratory work has begun and after preparing this work for several months we are all very excited to finally get it started. Today we started preparing sterile equipment and we also wrote risk declarations for all of our planned experiments. We also introduced ourselves and held a brief presentation of our project to the very nice people working at the department of System’s biology here at Chalmers University of Technology.Tuesday, 5th of June
Today our dear yeast strains that were kindly provided to us by the Kondo research group of Kobe University, Japan, were transferred to new and fresh YPD agar plates. We also prepared buffers needed for making competent E. coli and we inoculated E. coli DH5α in LB medium.
Wednesday, 6th of June
We prepared a stock of competent E. coli DH5α. Today we also amplified the shuttle plasmids pSP-GM1 and pIYC04 that we will use in our laboratory work, by transforming E. coli DH5α. The cells were then grown on LB-Amp agar plates. We also worked with the team wiki page and created a banner and a logo for the site.
Thursday, 7th of June
Not much laboratory could be done today. Two colonies from each plate containing the E. coli DH5α that were transformed with either pSP-GM1 or pIYC04, were inoculated in ampicillin-containing media and were grown overnight. For the rest of the day, we worked with creating a team brochure and we also established a Facebook page for the team. Click [http://www.facebook.com/ChalmersGothenburgiGEMTeam2012 here] to visit our page and Like our team!
Friday, 8th of June
We purified our pSP-GM1 and pIYC04 plasmids from the transformed E. coli.
 "
"