Team:Peking/Project
From 2012.igem.org
| (83 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
<div class="PKU_context floatR first"> | <div class="PKU_context floatR first"> | ||
| - | <h3 | + | <h3 id="title1">Synthetic Biology: What's Hindering Us?</h3> |
| - | <p> | + | <p>Efforts in synthetic biology have been primarily focused on the creation and perfection of biological modules and systems in order to achieve specific functions. Since first demonstrated by the bacterial toggle switch and the Repressilator in 2000, synthetic biologists now possess the ability to engineer and manipulate genetic circuits, metabolic pathways, and even entire genomes. <BR><BR> |
| - | + | In genetic engineering, chemicals are frequently employed as signals to control molecular and cellular behavior. However, because chemicals deliver information only by diffusion, chemical signals are seriously limited by the short-range interactions. Another serious issue regarding chemical signals is the complexity of the process in fully eradicating them, making it difficult for the system to reset. Additionally, the cytotoxicity, high cost, and narrow dynamic range of chemicals urge synthetic biologists to find more spatiotemporally specific and environmental friendly alternatives. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | + | <img src="/wiki/images/b/be/Peking2012_Chemical_Light_comparison.png" alt="comparison of Chemical and light" style="width:600px"/> | |
| - | + | <p class="description" style="text-align:center;font-style:normal;">Figure 1. Comparison of chemical signal and light signal</p> | |
| - | Figure | + | |
</div> | </div> | ||
| - | |||
| - | |||
</div> | </div> | ||
| - | |||
<div class="PKU_context floatR"> | <div class="PKU_context floatR"> | ||
| - | <h3 | + | <h3 id="title2">Optogenetics: Inspiring Synthetic Biology </h3> |
| - | <p> | + | <p>Literally, optogenetics refers to the genetic strategies which employ light as the input signal in the place of chemical signals. During the last decade, optogenetics has made significant impact on life sciences and other practices<sup><a href="#ref1" title=" Miesenbock, G. (2011). Optogenetic control of cells and circuits." Annu Rev Cell Dev Biol 27: 731-758.>[1]</a></sup>. It has evolved rapidly, with light-switchable promoters<sup><a href="#ref2" title=" Shimizu-Sato, S., E. Huq, et al. (2002). A light-switchable gene promoter system". Nat Biotechnol 20(10): 1041-1044.>[2]</a></sup>, light-induced protein-protein interaction<sup><a href="#ref3" title=" Levskaya, A., O. D. Weiner, et al. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction." Nature 461(7266): 997-1001.>[3]</a></sup>, light-activated ion channelsand<sup><a href="#ref4" title=" Zemelman, B. V., G. A. Lee, et al. (2002). Selective photostimulation of genetically chARGed neurons". Neuron 33(1): 15-22.>[4]</a></sup> eventually, light-controlled animal behavior, and so on. The spatiotemporal specificity of light signals allows for precise manipulation and long range interaction without physical contact. Besides its incredible range of functions, light resources are also cheaper, more sustainable, and environmentally friendly. These properties together make optogenetic tools attractive for synthetic biologists. |
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | < | + | |
| - | + | ||
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="" alt="[Fig 2.]" /> | + | <img src="/wiki/images/4/40/Peking2012_SA_Optogenetics.jpg" alt="[Fig 2.]" style="width:450px;"/> |
| - | <p class="description"> | + | <p class="description" style="text-align:center;font-style:normal;"> |
| - | + | Figure 2. Evolution of Optogenetics<sup><a href="#ref6">[6]</a><sup></p> | |
| + | <p>Light as a signal may carry massive information. The wavelength, intensity, frequency and spatial-temporal patterns of light are all informations encoded in a physical manner. Plus light signals possess a Bool-logic-like dynamic behavior and does not require any medium for its propagation, it would be much more enabling in transmitting accurate and complex information than the frequently investigated chemical signals. (In fact, natural biological systems has already explored this advantage to an astonishing extent. Think of our vision! ) <br/><br/> | ||
| + | |||
| + | Thus using light as a signal, we may program cellular behaviors to achieve, say, cell-cell communication that does not require physical contact. To communicate through light, first cells must be able to sense light, which means we need to develop a light sensor. And to realize this new generation of light communication that has never been implemented before, the sensor itself must make a big difference.</p> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| + | |||
<div class="PKU_context floatR"> | <div class="PKU_context floatR"> | ||
| - | <h3 class=" | + | <h3 id="title5">Reference</h3> |
| - | < | + | <p></p> |
| - | + | <ul class="refer"><li id="ref1"> | |
| - | </ | + | 1. Miesenbock, G. (2011) Optogenetic control of cells and circuits. <i>Annu. Rev. Cell. Dev. Biol.</i>, 27: 731: 758<br /> |
| + | </li><li id = "ref2"> | ||
| + | 2. Shimizu-Sato, S., Huq, E., <i>et al</i>. (2002) A light-switchable gene promoter system. <i>Nat. Biotechnol.</i>, 20(10): 1041: 1044<br /> | ||
| + | </li><li id = "ref3"> | ||
| + | 3. Levskaya, A., Weiner, O.D., <i>et al.</i> (2009) Spatiotemporal control of cell signalling using a light-switchable protein interaction. <i>Nature</i>, 461(7266): 997: 1001.<br /> | ||
| + | </li><li id = "ref4"> | ||
| + | 4. Zemelman, B.V., Lee, G.A., <i>et al.</i> (2002) Selective photostimulation of genetically chARGed neurons. <i>Neuron</i>, 33(1): 15: 22<br /> | ||
| + | </li><li id = "ref5"> | ||
| + | 5. Ohlendorf, R., Vidavski, R.R., <i>et al.</i> (2012) From dusk till dawn: one-plasmid systems for light-regulated gene expression. <i>J. Mol. Biol.</i> 416(4): 534: 542<br /> | ||
| + | </li><li id = "ref6"> | ||
| + | 6. http://www.stanford.edu/group/dlab/optogenetics/ | ||
| + | |||
| + | </li></ul> | ||
</div> | </div> | ||
| - | </html> | + | </html>{{Template:Peking2012_Color_Epilogue}} |
Latest revision as of 09:31, 26 October 2012
Synthetic Biology: What's Hindering Us?
Efforts in synthetic biology have been primarily focused on the creation and perfection of biological modules and systems in order to achieve specific functions. Since first demonstrated by the bacterial toggle switch and the Repressilator in 2000, synthetic biologists now possess the ability to engineer and manipulate genetic circuits, metabolic pathways, and even entire genomes.
In genetic engineering, chemicals are frequently employed as signals to control molecular and cellular behavior. However, because chemicals deliver information only by diffusion, chemical signals are seriously limited by the short-range interactions. Another serious issue regarding chemical signals is the complexity of the process in fully eradicating them, making it difficult for the system to reset. Additionally, the cytotoxicity, high cost, and narrow dynamic range of chemicals urge synthetic biologists to find more spatiotemporally specific and environmental friendly alternatives.
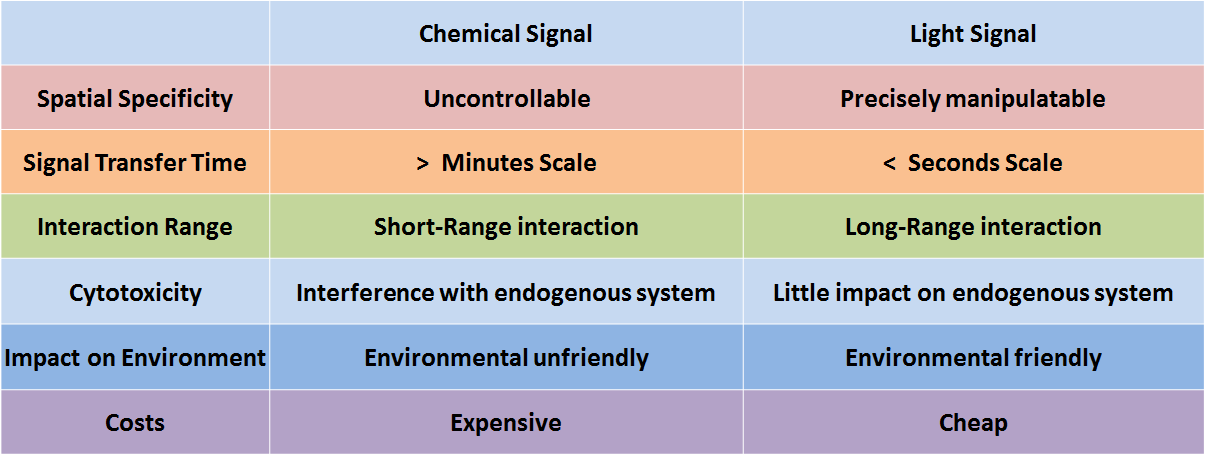
Figure 1. Comparison of chemical signal and light signal
Optogenetics: Inspiring Synthetic Biology
Literally, optogenetics refers to the genetic strategies which employ light as the input signal in the place of chemical signals. During the last decade, optogenetics has made significant impact on life sciences and other practices[1]. It has evolved rapidly, with light-switchable promoters[2], light-induced protein-protein interaction[3], light-activated ion channelsand[4] eventually, light-controlled animal behavior, and so on. The spatiotemporal specificity of light signals allows for precise manipulation and long range interaction without physical contact. Besides its incredible range of functions, light resources are also cheaper, more sustainable, and environmentally friendly. These properties together make optogenetic tools attractive for synthetic biologists.
![[Fig 2.]](/wiki/images/4/40/Peking2012_SA_Optogenetics.jpg)
Figure 2. Evolution of Optogenetics[6]
Light as a signal may carry massive information. The wavelength, intensity, frequency and spatial-temporal patterns of light are all informations encoded in a physical manner. Plus light signals possess a Bool-logic-like dynamic behavior and does not require any medium for its propagation, it would be much more enabling in transmitting accurate and complex information than the frequently investigated chemical signals. (In fact, natural biological systems has already explored this advantage to an astonishing extent. Think of our vision! )
Thus using light as a signal, we may program cellular behaviors to achieve, say, cell-cell communication that does not require physical contact. To communicate through light, first cells must be able to sense light, which means we need to develop a light sensor. And to realize this new generation of light communication that has never been implemented before, the sensor itself must make a big difference.
Reference
-
1. Miesenbock, G. (2011) Optogenetic control of cells and circuits. Annu. Rev. Cell. Dev. Biol., 27: 731: 758
-
2. Shimizu-Sato, S., Huq, E., et al. (2002) A light-switchable gene promoter system. Nat. Biotechnol., 20(10): 1041: 1044
-
3. Levskaya, A., Weiner, O.D., et al. (2009) Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature, 461(7266): 997: 1001.
-
4. Zemelman, B.V., Lee, G.A., et al. (2002) Selective photostimulation of genetically chARGed neurons. Neuron, 33(1): 15: 22
-
5. Ohlendorf, R., Vidavski, R.R., et al. (2012) From dusk till dawn: one-plasmid systems for light-regulated gene expression. J. Mol. Biol. 416(4): 534: 542
- 6. http://www.stanford.edu/group/dlab/optogenetics/
 "
"














