Team:Cambridge/Outreach/FutureDirections
From 2012.igem.org
(Created page with "=Chanel number six= One of the big features of our project is the use of a second fluorescence/ luminescence channel to give a reliable estimate of cell activity, against which ...") |
(→Potential applications) |
||
| (30 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Template:Team:Cambridge/CAM_2012_TEMPLATE_HEAD_OUTREACH}} | ||
| + | |||
=Chanel number six= | =Chanel number six= | ||
| + | |||
| + | [[File:ChanelNumberSixLogo.png|320px|right]] | ||
One of the big features of our project is the use of a second fluorescence/ luminescence channel to give a reliable estimate of cell activity, against which reporter activity can be adjusted. However, there is no need to limit ourselves to only two channels - the more channels that are used, the more data that can be squeezed out of a single biosensor strain. | One of the big features of our project is the use of a second fluorescence/ luminescence channel to give a reliable estimate of cell activity, against which reporter activity can be adjusted. However, there is no need to limit ourselves to only two channels - the more channels that are used, the more data that can be squeezed out of a single biosensor strain. | ||
| Line 6: | Line 10: | ||
An issue with this proposal is that with additional channels, there comes a greater risk of cross talk between the channels. This is part of the reason we have chosen to isolate our different biosensor strains in separate containers. In essence, each container is a different channel, separated in space rather than in wavelength. However, with the progression of our knowledge of the field of sythetic biology and the development of improved predictive software, we should be able to avoid this problem in the future. | An issue with this proposal is that with additional channels, there comes a greater risk of cross talk between the channels. This is part of the reason we have chosen to isolate our different biosensor strains in separate containers. In essence, each container is a different channel, separated in space rather than in wavelength. However, with the progression of our knowledge of the field of sythetic biology and the development of improved predictive software, we should be able to avoid this problem in the future. | ||
| + | |||
| + | [[File:QuestionableFPcurves.png|left|400px|thumb|One of the difficulties with using multiple channels. While this system would be fairly simple to deconvolve with only one additional sampling frequency, with multiple fluorescence channels uncertainties due to the unreliability of biological systems would add, resulting in highly unreliable readings.]] | ||
More fundamental is the problem of reliably distinguishing the different channels. While a two channel system is relatively simple to use, use of three or more fluorescent proteins may cause ambiguities in the data, as shown in the figure. These problems are similar to those seen in vision, where the overlap in the spectral sensitivities of the three cone types leads to impairment of our ability to accurately distinguish different hues. In this case though, the main difficulty is with the growth of unreliability of the measurements with increasing numbers of overlapping emission curves. | More fundamental is the problem of reliably distinguishing the different channels. While a two channel system is relatively simple to use, use of three or more fluorescent proteins may cause ambiguities in the data, as shown in the figure. These problems are similar to those seen in vision, where the overlap in the spectral sensitivities of the three cone types leads to impairment of our ability to accurately distinguish different hues. In this case though, the main difficulty is with the growth of unreliability of the measurements with increasing numbers of overlapping emission curves. | ||
| - | To get around this, we propose using additional ''forms'' of channel. For example, | + | |
| + | To get around this, we propose using additional ''forms'' of channel. For example, it may be possible to use the frequency of oscillation of a genetic circuit as an output. By using both amplitude and frequency of such an oscillatory system, it may be possible to encode two variables with a single fluorescent protein, effectively doubling the possible number of outputs. It is possible to concieve of other possible output channels - given that light is the most simple thing to detect in microbiological contexts (hence why fluorescent proteins and luminescent proteins are the most popular reporter choices for various assays of microscopic samples), it would be interesting to investigate the possibility of using other properties of the emitted light. Would it be possible to control the polorisation of the light, for example? Such a system would most likely need advances in cell fixation techniques, so as to ensure that the cells are aligned in the same direction. | ||
| + | |||
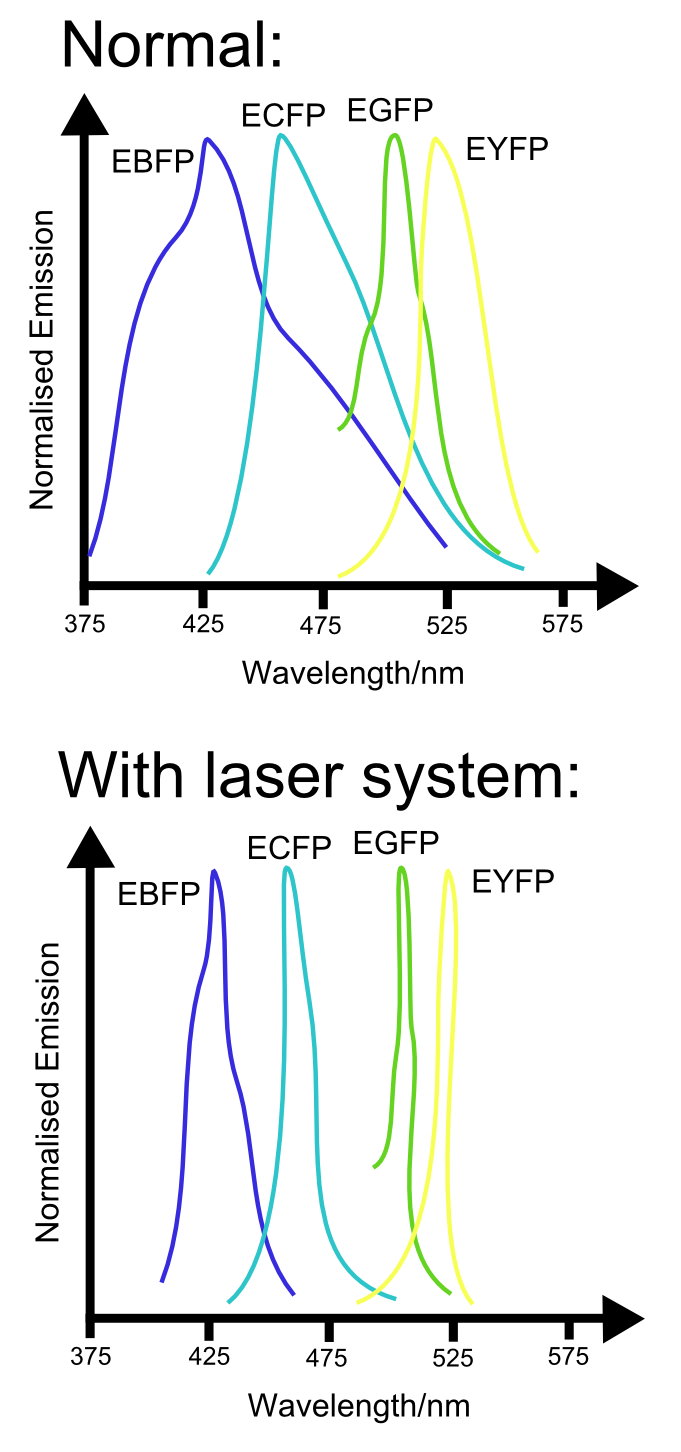
| + | [[File:LaserNormalFPComparison.png|right|300px|thumb|Comparison between normal emission spectrum and theoretical emission spectrum when using laser based system. Note the reduction in overlap between the different spectra, reducing the uncertainty in measurements taken at the spectral peaks.]] | ||
Alternatively, the development of techniques that enable GFP (and, in the future, most likely other fluorescent proteins) to act as lasers may sharpen the emission spectra of these proteins. In doing so, the number of different fluorescent proteins that could be sqeezed into the available spectral range would be greatly increaced. Again, this would enable us to use even more channels. | Alternatively, the development of techniques that enable GFP (and, in the future, most likely other fluorescent proteins) to act as lasers may sharpen the emission spectra of these proteins. In doing so, the number of different fluorescent proteins that could be sqeezed into the available spectral range would be greatly increaced. Again, this would enable us to use even more channels. | ||
Unfortuneately, such biological lasers have only been demonstrated in animal cells, and the cell walls of bacteria may interfere with the ability to generate effective lasing. Nevertheless, if a sensing standard within eukaryotic cells is ever developed, this may become a useful tool for increasing the range of such a system. | Unfortuneately, such biological lasers have only been demonstrated in animal cells, and the cell walls of bacteria may interfere with the ability to generate effective lasing. Nevertheless, if a sensing standard within eukaryotic cells is ever developed, this may become a useful tool for increasing the range of such a system. | ||
| - | |||
| - | |||
However, either of these techniques will require considerable leaps in our ability to create complex constructs. Gibson may be suitable, but from our experience it will need to be made more reliable before we use it to make the large constructions needed. | However, either of these techniques will require considerable leaps in our ability to create complex constructs. Gibson may be suitable, but from our experience it will need to be made more reliable before we use it to make the large constructions needed. | ||
| Line 21: | Line 28: | ||
==Potential applications== | ==Potential applications== | ||
| - | Should the prospect of reliable construction of plasmids with three or more channels become a reality, the oppertunities will be vast. One of the most simple (yet effective) may be the development of improved control systems for bioreactors. A three channel system should be all that is needed for the accurate and speedy detection of changes in the metabolism of cultured cells within the bioreactor. As discussed in this | + | Should the prospect of reliable construction of plasmids with three or more channels become a reality, the oppertunities will be vast. One of the most simple (yet effective) may be the development of improved control systems for bioreactors. A three channel system should be all that is needed for the accurate and speedy detection of changes in the metabolism of cultured cells within the bioreactor. As discussed in [http://schaechter.asmblog.org/schaechter/2009/11/the-limitations-of-lb-medium.html this article], magnesium may often becomes a limiting nutrient within bacterial cultures, causing a distinct shift in the metabolic properties of the cells. One channel would therefore be devoted to the measurement of magnesium in the culture with the [[Team:Cambridge/Project/MagnesiumRiboswitch|magnesium riboswitch]] we have characterized, causing automatic release of magnesium chloride by the control system into the medium. A second channel would monitor the levels of sigma factors usually only present during the plateau phase of growth, causing the release of fresh media if it detects that growth is about to slow down, and so prevent changes in the metabolic properties of the cells. The third channel would be made from a constitutive channel which would monitor the overall state of the cells, ensuring that their overall metabolism remained within acceptable bounds. |
| - | With increacing complexity of our ability to use multiple channels comes increacing complexity of the applications. | + | Such a strain would only need to be introduced into the bioreactor in small quantities, allowing the main, economically useful cells to dominate. Small samples would be continually taken, and the fluorescence channels quickly analysed to provide data for the control system. |
| + | |||
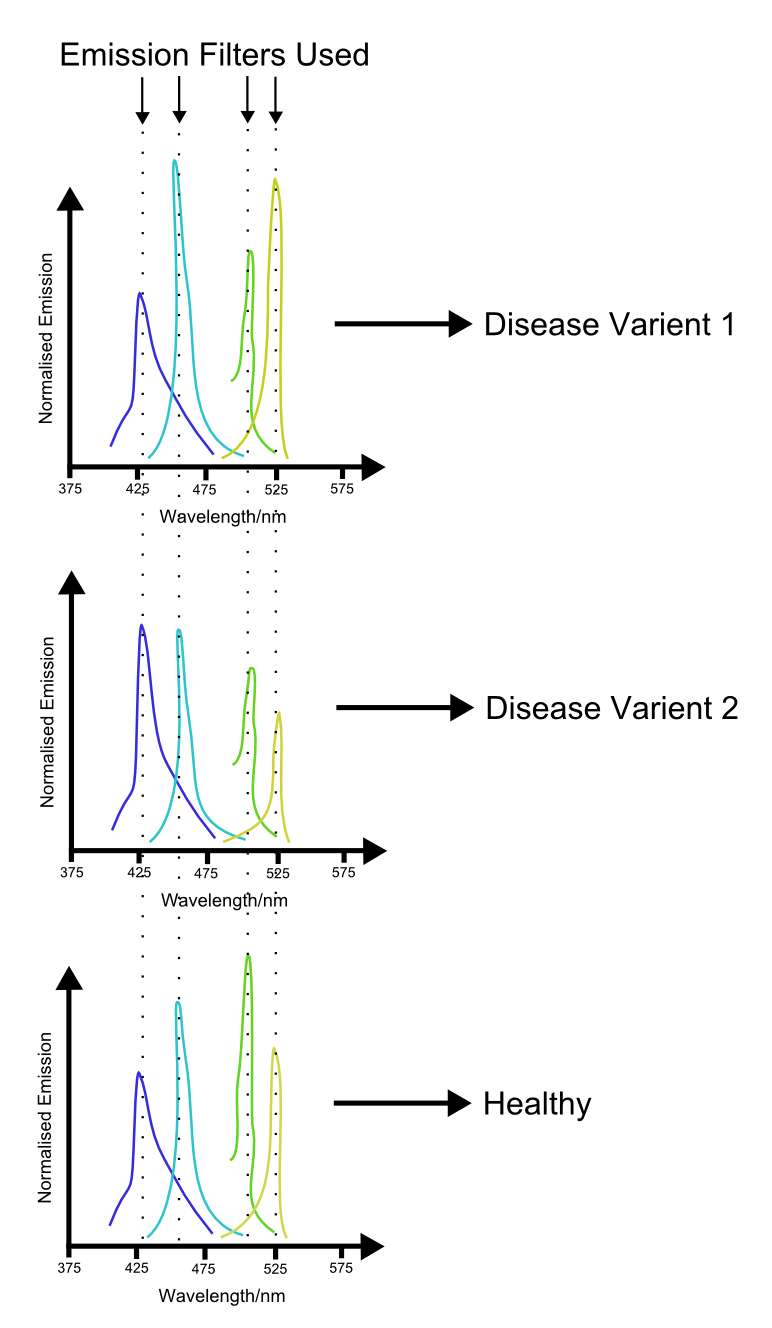
| + | [[File:FPDiseaseDiagnosis.png|500px|left|thumb|Possible way in which a multi-channel system may produce spectral 'fingerprints' to characterize different diseases.]] | ||
| + | |||
| + | With increacing complexity of our ability to use multiple channels comes increacing complexity of the applications. Further into the future, applications of such a system may include medical science. We propose that multiple indices of health be hooked up to different channels in such a system. Because the biosensing proteins involved in such a proposal are likely to only work in a eukaryotic environment, a eukaryotic chassis will need to be used. A blood sample from the patient is then innoculated with the sensory cells. An assay is performed by monitoring the output of the different channels, potentially using the laser system proposed above. The profile generated by this procedure may then produce a characteristic profile which can then be used to 'fingerprint' different diseases. | ||
| + | |||
| + | For example, a potent indicator of cardiovascular disease is the ratio of total cholesterol and HDL levels. [http://www.sciencemag.org/content/271/5248/518 HDL] and [http://www.sciencedirect.com/science/article/pii/0092867484901880 LDL] receptors exist which, while they do not directly couple to sensory pathways, could be adapted in the future to cause metabolic changes in a reporter cell. A three channel system, with total cholesterol, HDL levels and cell activity as its outputs would give a quick and single reaction assay for cardiovascular health. | ||
| + | |||
| + | Such a system, when combined with methods that are capable of analysing large quantities of data quickly, may also yield useful associations between many previously unchecked variables. With a construct with a series of steroid hormone detectors, for example, we would be able to discover the association between different disease types and different hormone levels. This data could then be put back into the diagnostic side of the system, improving the diagnostic power of the tool. | ||
| + | |||
| + | Extending this 'fingerprinting' idea into the far future, when GMOs have likely become a feature of everyday life, many interesting ideas can be thought of. For example, bacterial 'paints' could be developed which secreted different pigments according to the chemical profile of the underlying surface. After leaving to dry/develop for several hours, the specific chemical properties of the surface could be determined. This could be useful for geochemical surveys before well drilling, circumventing the problems that have been faced by such measures currently and, potentially, rendering our field testing kit obsolete. | ||
| + | |||
| + | Lastly, as the name of this section implies, we think an innovative product would be a perfume which responds to the wearers moods. Previous iGEM teams have already demonstrated the possibility of making e.coli more pleasingly fragrant, with the aromas of wintergreen and banana. Hooking up this output (a multichanel output in chemical space) to an output responsive to compounds in human sweat could, for example, cause the release of soothing eucalyptus oils in response to stress related compounds. How fragrant. | ||
===References=== | ===References=== | ||
*Gather M. and Yun S., Single-cell biological lasers, Nature Photonics (2011) '''5''', 406–410 | *Gather M. and Yun S., Single-cell biological lasers, Nature Photonics (2011) '''5''', 406–410 | ||
| + | *Prospective Studies Collaboration, Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths, The Lancet (2007), '''370''', Issue 9602, 1829–1839 | ||
| + | |||
| + | {{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | ||
Latest revision as of 23:01, 26 September 2012
Contents |
Chanel number six
One of the big features of our project is the use of a second fluorescence/ luminescence channel to give a reliable estimate of cell activity, against which reporter activity can be adjusted. However, there is no need to limit ourselves to only two channels - the more channels that are used, the more data that can be squeezed out of a single biosensor strain.
Design considerations
An issue with this proposal is that with additional channels, there comes a greater risk of cross talk between the channels. This is part of the reason we have chosen to isolate our different biosensor strains in separate containers. In essence, each container is a different channel, separated in space rather than in wavelength. However, with the progression of our knowledge of the field of sythetic biology and the development of improved predictive software, we should be able to avoid this problem in the future.
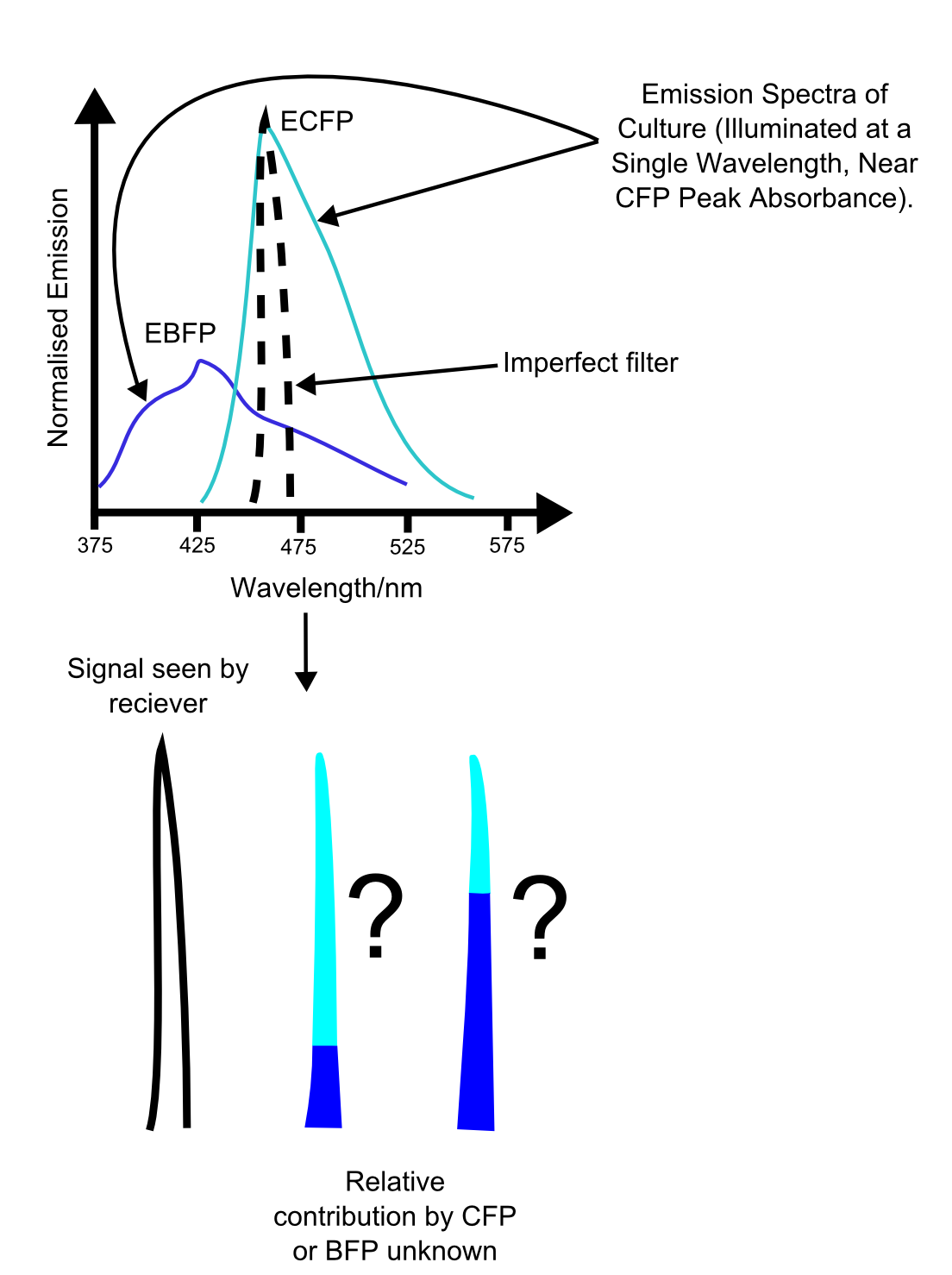
More fundamental is the problem of reliably distinguishing the different channels. While a two channel system is relatively simple to use, use of three or more fluorescent proteins may cause ambiguities in the data, as shown in the figure. These problems are similar to those seen in vision, where the overlap in the spectral sensitivities of the three cone types leads to impairment of our ability to accurately distinguish different hues. In this case though, the main difficulty is with the growth of unreliability of the measurements with increasing numbers of overlapping emission curves.
To get around this, we propose using additional forms of channel. For example, it may be possible to use the frequency of oscillation of a genetic circuit as an output. By using both amplitude and frequency of such an oscillatory system, it may be possible to encode two variables with a single fluorescent protein, effectively doubling the possible number of outputs. It is possible to concieve of other possible output channels - given that light is the most simple thing to detect in microbiological contexts (hence why fluorescent proteins and luminescent proteins are the most popular reporter choices for various assays of microscopic samples), it would be interesting to investigate the possibility of using other properties of the emitted light. Would it be possible to control the polorisation of the light, for example? Such a system would most likely need advances in cell fixation techniques, so as to ensure that the cells are aligned in the same direction.
Alternatively, the development of techniques that enable GFP (and, in the future, most likely other fluorescent proteins) to act as lasers may sharpen the emission spectra of these proteins. In doing so, the number of different fluorescent proteins that could be sqeezed into the available spectral range would be greatly increaced. Again, this would enable us to use even more channels.
Unfortuneately, such biological lasers have only been demonstrated in animal cells, and the cell walls of bacteria may interfere with the ability to generate effective lasing. Nevertheless, if a sensing standard within eukaryotic cells is ever developed, this may become a useful tool for increasing the range of such a system.
However, either of these techniques will require considerable leaps in our ability to create complex constructs. Gibson may be suitable, but from our experience it will need to be made more reliable before we use it to make the large constructions needed.
Potential applications
Should the prospect of reliable construction of plasmids with three or more channels become a reality, the oppertunities will be vast. One of the most simple (yet effective) may be the development of improved control systems for bioreactors. A three channel system should be all that is needed for the accurate and speedy detection of changes in the metabolism of cultured cells within the bioreactor. As discussed in [http://schaechter.asmblog.org/schaechter/2009/11/the-limitations-of-lb-medium.html this article], magnesium may often becomes a limiting nutrient within bacterial cultures, causing a distinct shift in the metabolic properties of the cells. One channel would therefore be devoted to the measurement of magnesium in the culture with the magnesium riboswitch we have characterized, causing automatic release of magnesium chloride by the control system into the medium. A second channel would monitor the levels of sigma factors usually only present during the plateau phase of growth, causing the release of fresh media if it detects that growth is about to slow down, and so prevent changes in the metabolic properties of the cells. The third channel would be made from a constitutive channel which would monitor the overall state of the cells, ensuring that their overall metabolism remained within acceptable bounds.
Such a strain would only need to be introduced into the bioreactor in small quantities, allowing the main, economically useful cells to dominate. Small samples would be continually taken, and the fluorescence channels quickly analysed to provide data for the control system.
With increacing complexity of our ability to use multiple channels comes increacing complexity of the applications. Further into the future, applications of such a system may include medical science. We propose that multiple indices of health be hooked up to different channels in such a system. Because the biosensing proteins involved in such a proposal are likely to only work in a eukaryotic environment, a eukaryotic chassis will need to be used. A blood sample from the patient is then innoculated with the sensory cells. An assay is performed by monitoring the output of the different channels, potentially using the laser system proposed above. The profile generated by this procedure may then produce a characteristic profile which can then be used to 'fingerprint' different diseases.
For example, a potent indicator of cardiovascular disease is the ratio of total cholesterol and HDL levels. [http://www.sciencemag.org/content/271/5248/518 HDL] and [http://www.sciencedirect.com/science/article/pii/0092867484901880 LDL] receptors exist which, while they do not directly couple to sensory pathways, could be adapted in the future to cause metabolic changes in a reporter cell. A three channel system, with total cholesterol, HDL levels and cell activity as its outputs would give a quick and single reaction assay for cardiovascular health.
Such a system, when combined with methods that are capable of analysing large quantities of data quickly, may also yield useful associations between many previously unchecked variables. With a construct with a series of steroid hormone detectors, for example, we would be able to discover the association between different disease types and different hormone levels. This data could then be put back into the diagnostic side of the system, improving the diagnostic power of the tool.
Extending this 'fingerprinting' idea into the far future, when GMOs have likely become a feature of everyday life, many interesting ideas can be thought of. For example, bacterial 'paints' could be developed which secreted different pigments according to the chemical profile of the underlying surface. After leaving to dry/develop for several hours, the specific chemical properties of the surface could be determined. This could be useful for geochemical surveys before well drilling, circumventing the problems that have been faced by such measures currently and, potentially, rendering our field testing kit obsolete.
Lastly, as the name of this section implies, we think an innovative product would be a perfume which responds to the wearers moods. Previous iGEM teams have already demonstrated the possibility of making e.coli more pleasingly fragrant, with the aromas of wintergreen and banana. Hooking up this output (a multichanel output in chemical space) to an output responsive to compounds in human sweat could, for example, cause the release of soothing eucalyptus oils in response to stress related compounds. How fragrant.
References
- Gather M. and Yun S., Single-cell biological lasers, Nature Photonics (2011) 5, 406–410
- Prospective Studies Collaboration, Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths, The Lancet (2007), 370, Issue 9602, 1829–1839
 "
"