Team:LMU-Munich/Data
From 2012.igem.org
| (66 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo2.jpg}} | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo2.jpg}} | ||
| + | [[File:Data banner.resized WORDS.JPG|620px|link=]] | ||
| - | == | + | [[Image:BacillusBioBrickBox.png|right|100px|link=https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks]] |
| + | <br> | ||
| + | <div class="box" style= "background-color:#e4f1d7"> | ||
| - | + | ==='''''Bacillus''B'''io'''B'''rick'''B'''ox=== | |
| - | + | ||
| - | + | </div> | |
| - | | | + | <div class="box"> |
| - | |< | + | ====Evaluation of ''B. subtilis'' vectors==== |
| - | | | + | {| "width=100%" style="text-align:center;"| |
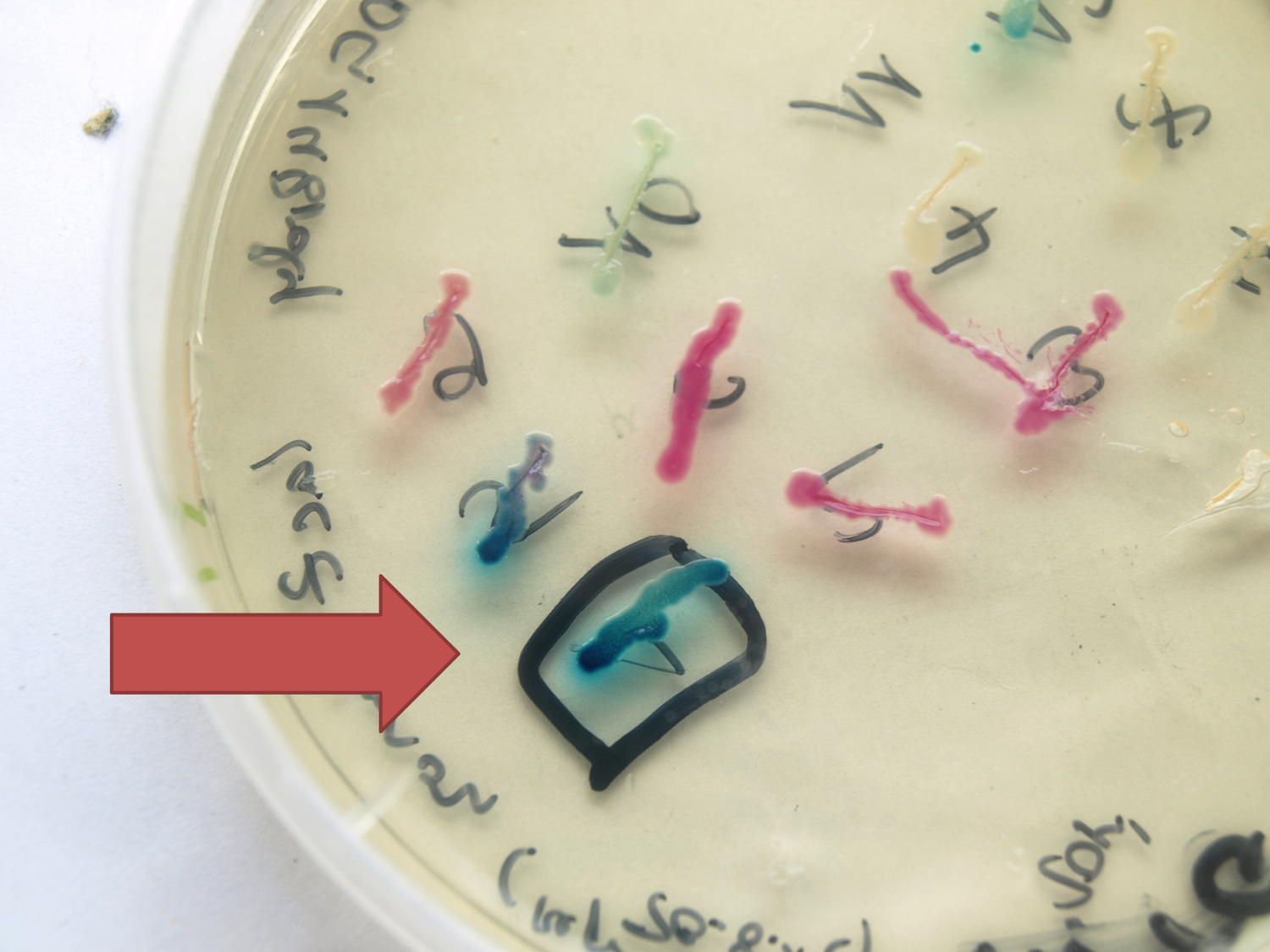
| + | |<p align="justify">Seven novel vectors were constructed and four already proven to work in ''B. subtilis''.</p> | ||
| + | |[[File:LacZ plate.png|right|100px|link=Team:LMU-Munich/Data/Vectors]] | ||
|- | |- | ||
| - | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Vectors]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| - | |||
<div class="box"> | <div class="box"> | ||
====Anderson Promoter Evaluation==== | ====Anderson Promoter Evaluation==== | ||
{| "width=100%" style="text-align:center;"| | {| "width=100%" style="text-align:center;"| | ||
| - | |<p align="justify"> | + | |<p align="justify">Eleven Anderson promoters were measured in ''B. subtilis'' and showed relatively weak activity.</p> |
|[[File:LMU Anderson preview.jpg|200px|link=Team:LMU-Munich/Data/Anderson]] | |[[File:LMU Anderson preview.jpg|200px|link=Team:LMU-Munich/Data/Anderson]] | ||
|- | |- | ||
| Line 69: | Line 31: | ||
====Constitutive ''Bacillus'' Promoters==== | ====Constitutive ''Bacillus'' Promoters==== | ||
{| "width=100%" style="text-align:center;"| | {| "width=100%" style="text-align:center;"| | ||
| - | |<p align="justify"> | + | |<p align="justify">Three novel ''B. subtilis'' promoter BioBricks (P<sub>''liaG''</sub>, P<sub>''veg''</sub>, P<sub>''lepA''</sub>) were constructed and functionality verified by luminescence and β-galactosidase assays.</p> |
|[[File:LMU Constitutive preview.jpg|200px|link=Team:LMU-Munich/Data/Constitutive]] | |[[File:LMU Constitutive preview.jpg|200px|link=Team:LMU-Munich/Data/Constitutive]] | ||
|- | |- | ||
| Line 78: | Line 40: | ||
====Inducible ''Bacillus'' Promoters==== | ====Inducible ''Bacillus'' Promoters==== | ||
{| "width=100%" style="text-align:center;"| | {| "width=100%" style="text-align:center;"| | ||
| - | |<p align="justify"> | + | |<p align="justify">A novel antibiotic-inducible promoter, P<sub>''liaI''</sub>, was constructed and thoroughly evaluated.</p> |
|[[File:Englisch Auswertung PliaI.png|200px|link=Team:LMU-Munich/Data/Inducible]] | |[[File:Englisch Auswertung PliaI.png|200px|link=Team:LMU-Munich/Data/Inducible]] | ||
|- | |- | ||
| Line 85: | Line 47: | ||
</div> | </div> | ||
| + | [[File:SporeCoat.png|90px|right|link=Team:LMU-Munich/Spore_Coat_Proteins]] | ||
| + | <br> | ||
<div class="box" style= "background-color:#e4f1d7"> | <div class="box" style= "background-color:#e4f1d7"> | ||
=='''Sporo'''beads== | =='''Sporo'''beads== | ||
| Line 90: | Line 54: | ||
<div class="box"> | <div class="box"> | ||
| - | === | + | ===Crust Promoter Evaluation=== |
{| "width=100%" style="text-align:center;"| | {| "width=100%" style="text-align:center;"| | ||
| - | |<p align="justify"> | + | |<p align="justify">Three sporulation-dependent promoters (P<sub>''cotYZ''</sub>, P<sub>''cotV''</sub>, P<sub>''cgeA''</sub>) were constructed, two of which show the expected behavior.</p> |
| - | |[[File: | + | |[[File:Promotoren.png|200px|link=Team:LMU-Munich/Data/crustpromoters]] |
|- | |- | ||
| - | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/ | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/crustpromoters]] |
|} | |} | ||
</div> | </div> | ||
| + | <div class="box"> | ||
| + | ===GFP-'''Sporo'''bead Evaluation=== | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify">They glow! GFP was successfully displayed on the spore crust as demonstrated by fluorescence microscopy.</p> | ||
| + | |[[File:Sporobead Constructs Dada sheet preview.png|200px|link=Team:LMU-Munich/Data/gfp_spore]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/gfp_spore]] | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | ==='''Sporo'''bead Purification=== | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify">We compared different ways of purifying ''Bacillus'' spores from vegetative cells: Lysozyme is the best!</p> | ||
| + | |[[File:Purification preview.png|200px|link=Team:LMU-Munich/Data/Sporepurification]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Sporepurification]] | ||
| + | |} | ||
| + | </div> | ||
| - | |||
| - | < | + | [[File:GerminationSTOP.png|90px|right|link=Team:LMU-Munich/Germination_Stop]] |
| + | <br> | ||
| + | <div class="box" style= "background-color:#e4f1d7"> | ||
| + | ==='''Germination'''STOP=== | ||
| + | </div> | ||
| - | + | <div class="box"> | |
| + | ===Knockouts of germination genes === | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify">We successfully prevented spore germination by multiple gene knockouts.</p> | ||
| + | |[[File:LMU Knockout previewii.jpg|200px|link=Team:LMU-Munich/Data/Knockout]] | ||
|- | |- | ||
| - | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Knockout]] | |
| - | + | |} | |
| - | | | + | </div> |
| - | + | <div class="box"> | |
| - | + | ==='''Suicide'''Switch === | |
| - | + | {| "width=100%" style="text-align:center;"| | |
| - | + | |<p align="justify">Death upon germination: We developed a novel safety strategy as a back-up to the gene knockouts.</p> | |
| - | + | |[[File:PlatereaderSuicideSwitch1.jpg|200px|link=Team:LMU-Munich/Data/Suicideswitch]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Suicideswitch]] | ||
|} | |} | ||
| + | </div> | ||
| - | < | + | [[File:LMU-Munich-Invertersign.png|90px|right|link=Team:LMU-Munich/Inverter]] |
| + | <br> | ||
| + | <div class="box" style= "background-color:#e4f1d7"> | ||
| + | ===Inverter=== | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | ===β-galactosidase assay of Inverter with ''lacZα'' === | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify">It works! Invert the output of your promoter of choice.</p> | ||
| + | |[[File:LMU Inverter graph.png|200px|link=Team:LMU-Munich/Data/Inverter]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Inverter]] | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="box" style= "background-color:#e4f1d7"> | ||
| + | ==='''Trash'''can=== | ||
| + | Things that were a pain in the ''neck''! | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | ===''cge''A and P<sub>''cge''A</sub>=== | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify"> | ||
| + | Unexpected restriction sites combined with no activity at all - what a bummer!</p> | ||
| + | |[[File:LMU cgea preview.jpg|200px|link=Team:LMU-Munich/Data/cgeA]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/cgeA]] | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | ===MazF=== | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify"> | ||
| + | The story of trying to clone a toxin from ''E. coli'' in ''E. coli'': They die, dammit! (...not a surprise, really...) | ||
| + | </p> | ||
| + | |[[File:MazFtrashcan.jpg|200px|link=Team:LMU-Munich/Data/MazF]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/MazF]] | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | ===Xylose promoters=== | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify"> | ||
| + | All constructs involving xylose-inducible promoters refused cloning or evaluation (sorry, Groningen...!). | ||
| + | </p> | ||
| + | |[[File:100px-D-Xylose Keilstrich.png|100px|link=Team:LMU-Munich/Data/Pxyl]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Pxyl]] | ||
| + | |} | ||
| + | </div> | ||
| + | <p></p> | ||
| Line 178: | Line 178: | ||
| + | For our big breakthroughs, or to follow specific projects, see the individual project pages: | ||
| - | + | <div class="box"> | |
| - | + | ====Project Navigation==== | |
| - | + | {| width="100%" align="center" style="text-align:center;" | |
| - | + | |[[File:Bacilluss_Intro.png|100px|link=Team:LMU-Munich/Bacillus_Introduction]] | |
| - | + | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] | |
| - | === | + | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] |
| - | + | |[[File:GerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] | |
| - | + | |- | |
| - | [[File: | + | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] |
| - | + | |[[Team:LMU-Munich/Bacillus_BioBricks|<font size="2" face="verdana">'''''Bacillus'''''<BR>'''B'''io'''B'''rick'''B'''ox</font>]] | |
| - | + | |[[Team:LMU-Munich/Spore_Coat_Proteins|<font size="2" face="verdana">'''Sporo'''beads</font>]] | |
| + | |[[Team:LMU-Munich/Germination_Stop|<font size="2" face="verdana">'''Germination'''<BR>STOP</font>]] | ||
| + | |} | ||
| + | </div> | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 14:31, 22 October 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

BacillusBioBrickBox
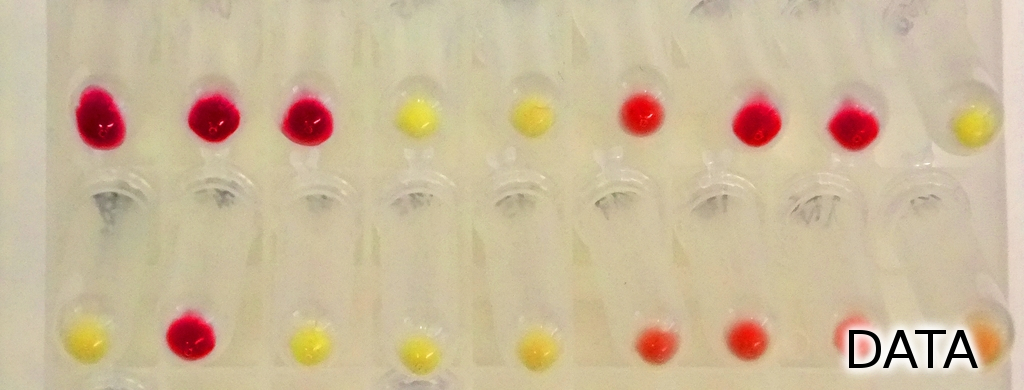
Evaluation of B. subtilis vectors
Seven novel vectors were constructed and four already proven to work in B. subtilis. | |
Anderson Promoter Evaluation
Eleven Anderson promoters were measured in B. subtilis and showed relatively weak activity. | 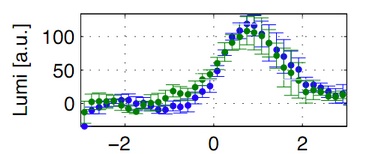
|
Constitutive Bacillus Promoters
Three novel B. subtilis promoter BioBricks (PliaG, Pveg, PlepA) were constructed and functionality verified by luminescence and β-galactosidase assays. | 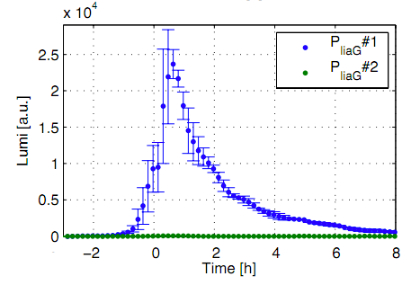
|
Inducible Bacillus Promoters
A novel antibiotic-inducible promoter, PliaI, was constructed and thoroughly evaluated. | 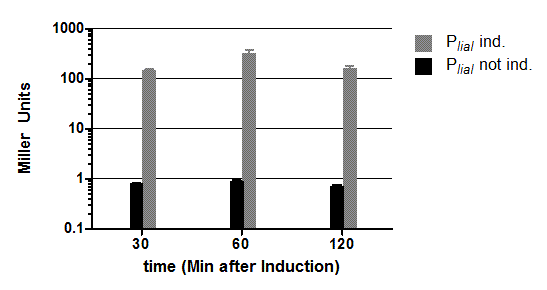
|
Sporobeads
Crust Promoter Evaluation
Three sporulation-dependent promoters (PcotYZ, PcotV, PcgeA) were constructed, two of which show the expected behavior. | 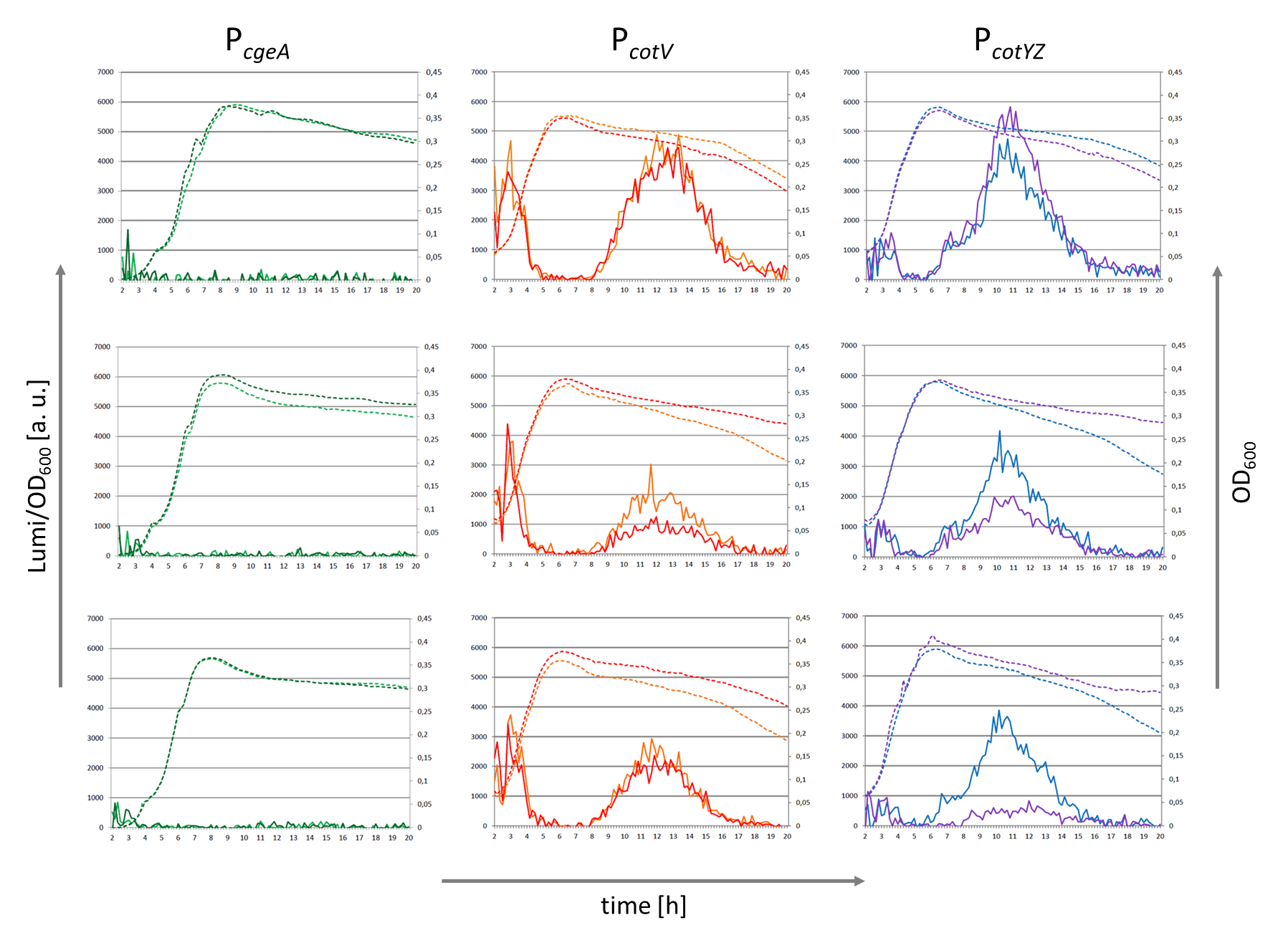
|
GFP-Sporobead Evaluation
They glow! GFP was successfully displayed on the spore crust as demonstrated by fluorescence microscopy. | 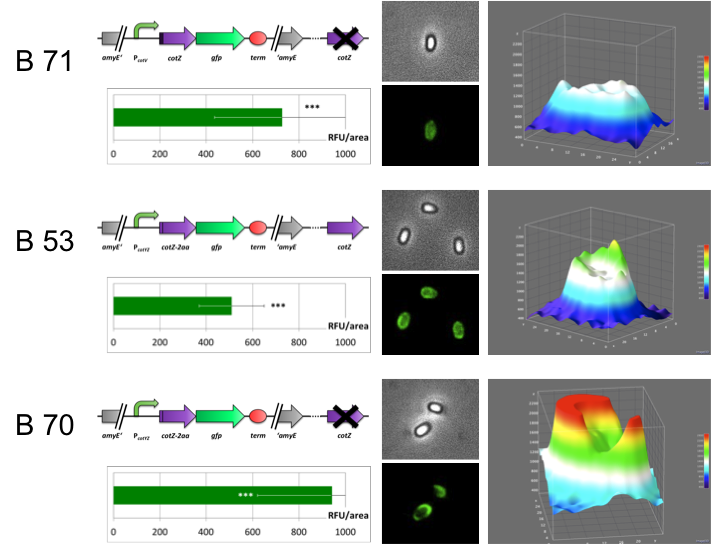
|
Sporobead Purification
We compared different ways of purifying Bacillus spores from vegetative cells: Lysozyme is the best! | 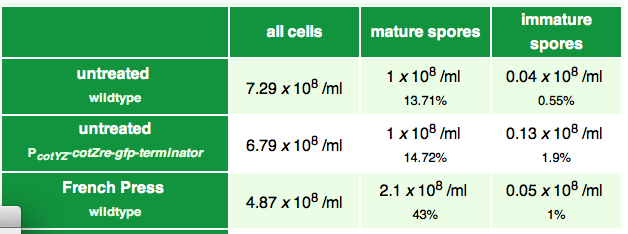
|
GerminationSTOP
Knockouts of germination genes
We successfully prevented spore germination by multiple gene knockouts. | 
|
SuicideSwitch
Death upon germination: We developed a novel safety strategy as a back-up to the gene knockouts. | 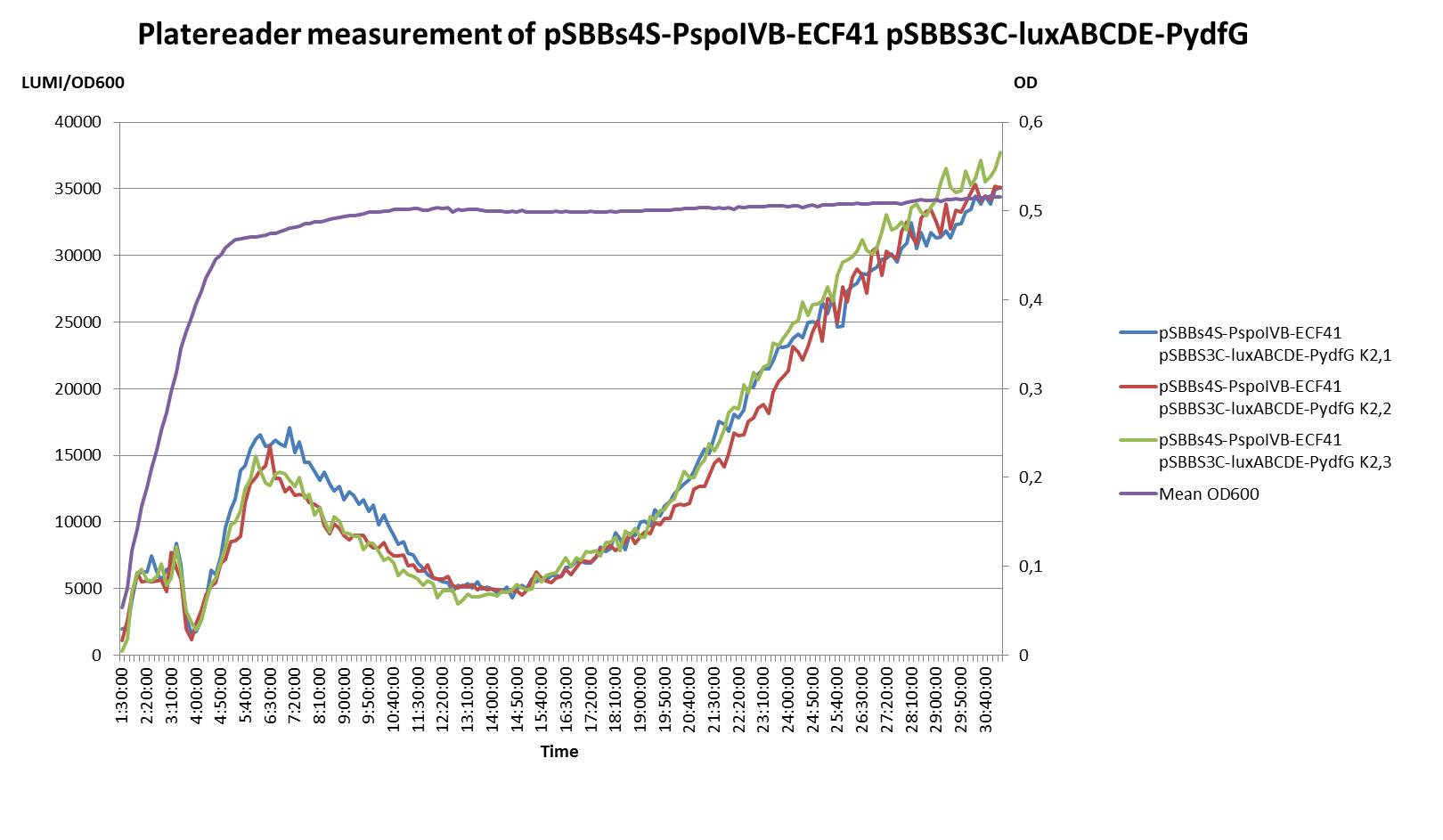
|
Inverter
β-galactosidase assay of Inverter with lacZα
It works! Invert the output of your promoter of choice. | 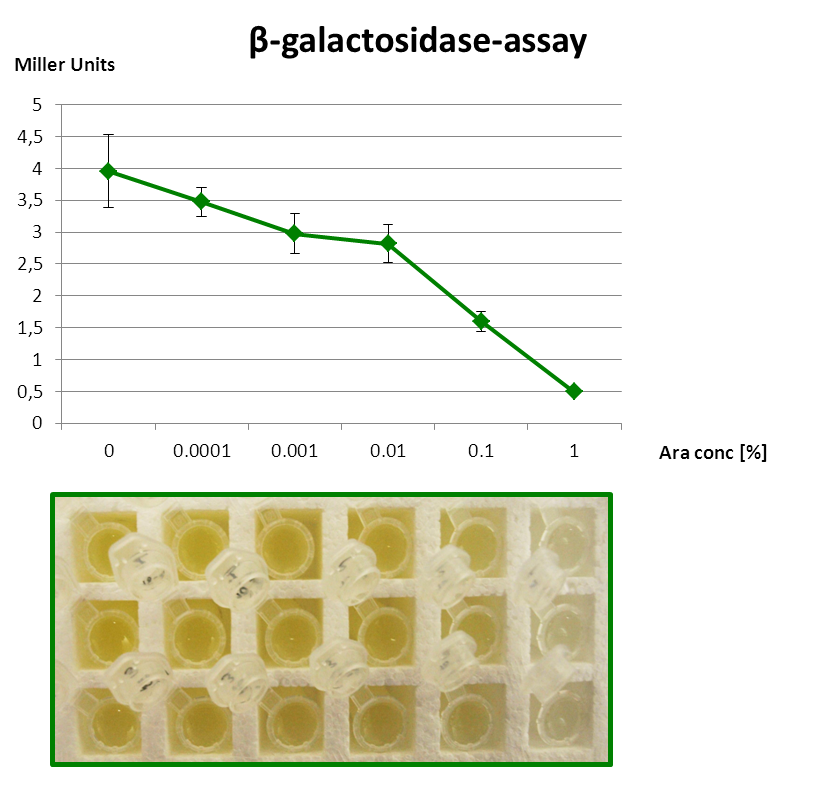
|
Trashcan
Things that were a pain in the neck!
MazF
The story of trying to clone a toxin from E. coli in E. coli: They die, dammit! (...not a surprise, really...) | 
|
Xylose promoters
All constructs involving xylose-inducible promoters refused cloning or evaluation (sorry, Groningen...!). | 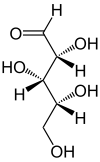
|
For our big breakthroughs, or to follow specific projects, see the individual project pages:
 "
"