Team:LMU-Munich/Test
From 2012.igem.org
| (48 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!-- Include the next line at the beginning of every page --> | <!-- Include the next line at the beginning of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo1.jpg}} | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo1.jpg}} | ||
| + | |||
| + | ==Our Team== | ||
| + | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 100%;background-color: #EBFCE4;" | | ||
| + | {| style="width: 100%" align="center" | ||
| + | |[[Image:IGEM2012_LMU_team_Korinna.jpg|Korinna|150px|center]] | ||
| + | |[[Image:IGEM2012_LMU_team_Franzi.jpg|Franzi|150px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Tamara.jpg|Tamara|150px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Simon.jpg|Simon|150px]] | ||
| + | |- | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Korinna | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Franzi | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Tamara | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Simon | ||
| + | |} | ||
| + | |- | ||
| + | | style="width: 100%;background-color: #EBFCE4;" | | ||
| + | {| style="width: 100%" align="center" | ||
| + | |[[Image:IGEM2012_LMU_team_Julia.jpg|150px|center]] | ||
| + | |[[Image:IGEM2012_LMU_team_Jenny.jpg|150px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Jara.jpg|150px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Thorsten.jpg|150px]] | ||
| + | |- | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Julia | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Jenny | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Jara | ||
| + | | style="background-color:#EBFCE4;text-align:center;" | | ||
| + | Thorsten | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Korinna</font> | ||
| + | |} | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Franzi</font> | ||
| + | |} | ||
| + | |- | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | Image:IGEM2012_LMU_team_Korinna.jpg|Korinna | ||
| + | Image:IGEM2012_LMU_team_Franzi.jpg|Franzi | ||
| + | Image:IGEM2012_LMU_team_Tamara.jpg|Tamara | ||
| + | Image:IGEM2012_LMU_team_Simon.jpg|Simon | ||
| + | Image:IGEM2012_LMU_team_Julia.jpg|Julia | ||
| + | Image:IGEM2012_LMU_team_Jenny.jpg|Jenny | ||
| + | Image:IGEM2012_LMU_team_Jara.jpg|Jara | ||
| + | Image:IGEM2012_LMU_team_Thorsten.jpg|Prof. Dr. Mascher | ||
| + | |||
| + | |[[Image:IGEM2012_LMU_team_Julia.jpg|Julia|100px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Jenny.jpg|Jenny|100px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Jara.jpg|Jara|100px]] | ||
| + | |[[Image:IGEM2012_LMU_team_Thorsten.jpg|Prof. Dr. Mascher|100px]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
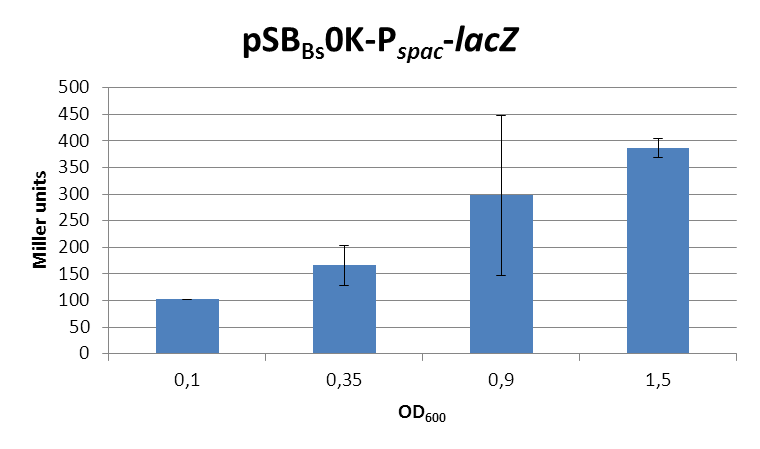
| + | <font color="#000000"; size="2">''lacZ''-activity in Miller units in pSB<sub>Bs</sub>0K-P<sub>''spac''</sub> depending on ''B. subtilis'' growth phase.</font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:LMU-Munich-0K-OD.png|500px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">''lacZ''-activity in Miller units in pSB<sub>Bs</sub>0K-P<sub>''spac''</sub> depending on ''B. subtilis'' growth phase.</font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | We also tested the [http://partsregistry.org/Promoters/Catalog/Anderson Anderson promoter collection] in ''Bacillus subtilis'' with pSB<sub>Bs</sub>3C-<i>luxABCDE</i>. The data will be online in our [https://2012.igem.org/Team:LMU-Munich/Data Data] section soon.</p> | ||
| + | <p align="justify">Furthermore we plan to submit reporter genes optimized for ''Bacillus subtilis'', namely ''GFP'', ''lacZ'', ''luc'' and ''mKate2''. </p> | ||
| + | Useful for the registry are also 5 tags in Freiburg Standard (cMyc, 10His, Flag, Strep, HA). | ||
| + | More details to come soon. | ||
| + | |||
<div class="box"> | <div class="box"> | ||
| - | {| style=" | + | ====Project Navigation==== |
| + | {| width="100%" align="center" style="text-align:center;" | ||
| + | |[[File:Bacilluss_Intro.png|100px|link=Team:LMU-Munich/Bacillus_Introduction]] | ||
| + | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] | ||
| + | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] | ||
| + | |[[File:GerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] | ||
| + | |- | ||
| + | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] | ||
| + | |[[Team:LMU-Munich/Bacillus_BioBricks|<font size="2" face="verdana">'''''Bacillus'''''<BR>'''B'''io'''B'''rick'''B'''ox</font>]] | ||
| + | |[[Team:LMU-Munich/Spore_Coat_Proteins|<font size="2" face="verdana">'''Sporo'''beads</font>]] | ||
| + | |[[Team:LMU-Munich/Germination_Stop|<font size="2" face="verdana">'''Germination'''<BR>STOP</font>]] | ||
| + | |} | ||
| + | </div> | ||
| - | + | ||
| - | ''' | + | <div class="box"> |
| - | | style="width | + | '''Project Navigation''' |
| - | + | {| style= cellpadding="10" width="100%" cellspacing="1" align="center" style="text-align:center;" | |
| - | | style=" | + | |style="background-color:white"| |
| - | + | {| | |
| - | | style=" | + | |style= "background-color:white"| |
| - | + | [[File:Bacilluss_Intro.png|120px|link=https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction]] | |
| + | |style= "background-color:white"| | ||
| + | [[Team:LMU-Munich/Bacillus_Introduction|''B. subtilis'' Intro]] | ||
|- | |- | ||
| - | | | + | |style= "background-color:white"| |
| - | + | [[File:BacillusBioBrickBox.png|120px|link=https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks]] | |
| - | | | + | |style= "background-color:white"| |
| - | + | [[Team:LMU-Munich/Bacillus_BioBricks|'''''Bacillus''B'''io'''B'''rick'''B'''ox]] | |
|- | |- | ||
| - | | | + | |} |
| + | |style="background-color:white"| | ||
| + | {| | ||
| + | |style="background-color:white"| | ||
| + | [[File:SporeCoat.png|100px|link=https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins]] | ||
| + | |style="background-color:white"| | ||
| + | [[Team:LMU-Munich/Spore_Coat_Proteins|'''Sporo'''beads]] | ||
| + | |- | ||
| + | |style="background-color:white"| | ||
| + | [[File:GerminationSTOP.png|90px|link=https://2012.igem.org/Team:LMU-Munich/Germination_Stop]] | ||
| + | |style="background-color:white"| | ||
| + | [[Team:LMU-Munich/Germination_Stop|'''Germination'''STOP]] | ||
| + | |} | ||
| + | |} | ||
| + | </div> | ||
| - | |||
| - | + | {| | |
| + | | <html><a href="https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=50"/></a></html> | ||
| - | pSB<sub>Bs</sub>3C | + | | [[Team:LMU-Munich/Bacillus_Introduction|Intro to ''Bacillus subtilis'']] |
| - | |Pveg | + | |
| + | |- | ||
| + | |||
| + | | <html><a href="https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=50"/></a></html> | ||
| + | |||
| + | |[[Team:LMU-Munich/Bacillus_BioBricks|''Bacillus'' BioBrick'''BOX''']] | ||
| + | |||
| + | |- | ||
| + | |||
| + | |<html><a href="https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=50"/></a></html> | ||
| + | |||
| + | |[[Team:LMU-Munich/Spore_Coat_Proteins|Sporo'''beads''']] | ||
| + | |- | ||
| + | |||
| + | |<html><a href="https://2012.igem.org/Team:LMU-Munich/Germination_Stop"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=50"/></a></html> | ||
| + | |||
| + | |[[Team:LMU-Munich/Germination_Stop|Germination'''STOP''']] | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | The promoters we submit are: | ||
| + | |||
| + | Pveg, PliaI, PliaG, plepA, Pxyl, Pxyl+XylR | ||
| + | |||
| + | <p align="justify">We also tested the [http://partsregistry.org/Promoters/Catalog/Anderson Anderson promoter collection] in ''Bacillus subtilis'' with pSB<sub>Bs</sub>3C-<i>luxABCDE</i>. The data will be online in our [https://2012.igem.org/Team:LMU-Munich/Data Data] section soon.</p> | ||
| + | |||
| + | <p align="justify">Furthermore we plan to submit reporter genes optimized for ''Bacillus subtilis'', namely ''GFP'', ''lacZ'', ''luc'' and ''mKate2''. </p> | ||
| + | |||
| + | Useful for the registry are also 5 tags in Freiburg Standard (cMyc, 10His, Flag, Strep, HA). | ||
| + | |||
| + | More details to come soon. | ||
| + | |||
| + | {| width="100%" | ||
| + | |valign="top" style="text-align:center;"|<font size="2"> | ||
| + | pSB<sub>Bs</sub>1C | ||
| + | |||
| + | pSB<sub>Bs</sub>4S | ||
| + | |||
| + | pSB<sub>Bs</sub>2E | ||
| + | |||
| + | pSB<sub>Bs</sub>1C-''lac''Z | ||
| + | |||
| + | pSB<sub>Bs</sub>3C-''lux''ABCDE | ||
| + | |||
| + | pSB<sub>Bs</sub>4S-P<sub>''Xyl''</sub> | ||
| + | |||
| + | pSB<sub>Bs</sub>0K-P<sub>''spac''</sub> | ||
| + | </font> | ||
| + | |valign="top"|<font size="2"> | ||
| + | Pveg | ||
PliaI | PliaI | ||
| Line 33: | Line 209: | ||
Anderson | Anderson | ||
| - | | | + | |
| + | </font> | ||
| + | |valign="top"|<font size="2"> | ||
mkate2 | mkate2 | ||
| Line 39: | Line 217: | ||
lacZ | lacZ | ||
| - | |||
| + | </font> | ||
| + | |valign="top"|<font size="2"> | ||
Flag | Flag | ||
| Line 46: | Line 225: | ||
cMyc | cMyc | ||
| + | |||
| + | Strep | ||
| + | |||
| + | HA | ||
| + | </font> | ||
|- | |- | ||
|} | |} | ||
| - | + | ||
<div class="box" style= "background-color:#e4f1d7"> | <div class="box" style= "background-color:#e4f1d7"> | ||
| Line 59: | Line 243: | ||
|<p align="justify">Evaluation of the Anderson library in ''B. subtilis'' with pSB<sub>Bs</sub>3C-''lux'' (luminescence) and pSB<sub>Bs</sub>1C-''lacZ'' | |<p align="justify">Evaluation of the Anderson library in ''B. subtilis'' with pSB<sub>Bs</sub>3C-''lux'' (luminescence) and pSB<sub>Bs</sub>1C-''lacZ'' | ||
[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Anderson]]</p> | [[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/Anderson]]</p> | ||
| + | |[[File:LMU Anderson preview.jpg|200px|link=Team:LMU-Munich/Data/Anderson]] | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | <div class="box"> | ||
| + | ====Anderson Promoter Evaluation==== | ||
| + | {| "width=100%" style="text-align:center;"| | ||
| + | |<p align="justify">Evaluation of the Anderson library in ''B. subtilis'' with pSB<sub>Bs</sub>3C-''lux'' (luminescence) and pSB<sub>Bs</sub>1C-''lacZ''</p> | ||
|[[File:LMU Anderson preview.jpg|200px|link=Team:LMU-Munich/Data/Anderson]] | |[[File:LMU Anderson preview.jpg|200px|link=Team:LMU-Munich/Data/Anderson]] | ||
|- | |- | ||
| Line 64: | Line 256: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
| - | + | <div class="box"> | |
| - | | style="width: | + | {| width="100%" |
| - | + | ||
| - | |[[ | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]]''' |
| - | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Promoters|Promoters]]''' | |
| + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Reporters|Reporters]]''' | ||
| + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks#Affinity_Tags|Affinity tags]] | ||
|- | |- | ||
| - | | | + | |[[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]] |
| + | |[[File:LMU PromoterIconBC.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Promoters]] | ||
| + | |[[File:LMU Reporter.png|60px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Reporters]] | ||
| + | |[[File:Proteinaffinitytagbutton.png|50px|link=Team:LMU-Munich/Bacillus_BioBricks#Affinity_Tags]] | ||
|- | |- | ||
|} | |} | ||
| + | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<!-- Include the next line at the end of every page --> | <!-- Include the next line at the end of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 22:02, 25 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Our Team
| ||||||||
|
| style="width: 70%;background-color: #EBFCE4;" |
|
Korinna |
| style="width: 70%;background-color: #EBFCE4;" |
|
Franzi |
|- |} |}
Image:IGEM2012_LMU_team_Korinna.jpg|Korinna Image:IGEM2012_LMU_team_Franzi.jpg|Franzi Image:IGEM2012_LMU_team_Tamara.jpg|Tamara Image:IGEM2012_LMU_team_Simon.jpg|Simon Image:IGEM2012_LMU_team_Julia.jpg|Julia Image:IGEM2012_LMU_team_Jenny.jpg|Jenny Image:IGEM2012_LMU_team_Jara.jpg|Jara Image:IGEM2012_LMU_team_Thorsten.jpg|Prof. Dr. Mascher
| |
| |
| |
| |-
| style="width: 70%;background-color: #EBFCE4;" |
|-
| style="width: 70%;background-color: #EBFCE4;" |
|
lacZ-activity in Miller units in pSBBs0K-Pspac depending on B. subtilis growth phase. |
|} |}
|
We also tested the [http://partsregistry.org/Promoters/Catalog/Anderson Anderson promoter collection] in Bacillus subtilis with pSBBs3C-luxABCDE. The data will be online in our Data section soon.</p>
Furthermore we plan to submit reporter genes optimized for Bacillus subtilis, namely GFP, lacZ, luc and mKate2.
Useful for the registry are also 5 tags in Freiburg Standard (cMyc, 10His, Flag, Strep, HA). More details to come soon.
Project Navigation
|
|
|

| Intro to Bacillus subtilis |

| Bacillus BioBrickBOX |

| Sporobeads |

| GerminationSTOP |
The promoters we submit are:
Pveg, PliaI, PliaG, plepA, Pxyl, Pxyl+XylR
We also tested the [http://partsregistry.org/Promoters/Catalog/Anderson Anderson promoter collection] in Bacillus subtilis with pSBBs3C-luxABCDE. The data will be online in our Data section soon.
Furthermore we plan to submit reporter genes optimized for Bacillus subtilis, namely GFP, lacZ, luc and mKate2.
Useful for the registry are also 5 tags in Freiburg Standard (cMyc, 10His, Flag, Strep, HA).
More details to come soon.
|
pSBBs1C pSBBs4S pSBBs2E pSBBs1C-lacZ pSBBs3C-luxABCDE pSBBs4S-PXyl pSBBs0K-Pspac |
Pveg PliaI PlepA Anderson |
mkate2 luc lacZ |
Flag His cMyc Strep HA |
BacillusBioBrickBox
Anderson Promoter Evaluation
Evaluation of the Anderson library in B. subtilis with pSBBs3C-lux (luminescence) and pSBBs1C-lacZ
| 
|
Anderson Promoter Evaluation
Evaluation of the Anderson library in B. subtilis with pSBBs3C-lux (luminescence) and pSBBs1C-lacZ | 
|
 "
"