Team:Wageningen UR/Journal/week16
From 2012.igem.org
(→Lab work) |
(→Lab work) |
||
| (8 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
== Lab work == | == Lab work == | ||
| - | New competent cell JM-109 was made this week, transformation efficiency was checked, which was 1*10^6. | + | New competent cell used for protein expression (JM-109) was made this week, transformation efficiency was checked, which was 1*10^6. |
| + | |||
| + | |||
| + | ''' CCMV ''' | ||
| + | |||
| + | ''13th August'' (Hugo) | ||
| + | <br/> | ||
| + | Colony PCR on IPTG_CCMV_NEG transformant colonies and 5 red BBA_J3901 colonies. | ||
| + | <br/> | ||
| + | Grew two colonies of IPTG_CCMV_NEG overnight for miniprepping. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ''14th August'' (Hugo) | ||
| + | <br/> | ||
| + | Robert miniprepped the overnight cultures that were inoculated at 13-08. Then I ran a digestion check with EcoRI + PstI. | ||
| + | <br/><br/> | ||
| + | Sent sample with CCMV neg in the chloramphenicol backbone for sequencing (GATC). | ||
| + | <br/> | ||
| + | Prepared iron-free E. coli medium. | ||
| + | <br/> | ||
| + | ''16 & 17th August'' (Hugo) | ||
| + | <br/> | ||
| + | Prepared JM 109 competent cells. | ||
| + | <br/><br/> | ||
| + | |||
| + | |||
| + | |||
'''TuYV''' | '''TuYV''' | ||
| - | * Last week's 'brick' minipreps were PCR checked for the correct insert. Results were unclear at first, but a second check gave positive results for the TuYV Coat Protein brick (1), the CP + Histidine tag brick(1H), and the CP + readthrough brick (4)! Only | + | * Last week's 'brick' minipreps were PCR checked for the correct insert. Results were unclear at first, but a second check gave positive results for the TuYV Coat Protein brick (1), the CP + Histidine tag brick(1H), and the CP + readthrough brick (4)! Only the CP + readthrough + His-tag(4H) brick wasn't found. |
| - | * The second trial of putting TuYV | + | * The second trial of putting TuYV constructs after IPTG induced promoter turned out to be a failure. |
| Line 59: | Line 85: | ||
10: 4˚C; ∞ | 10: 4˚C; ∞ | ||
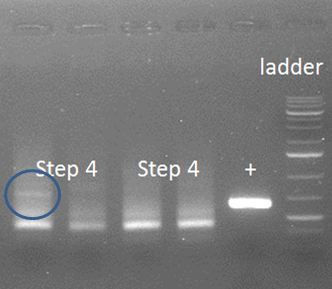
| - | [[File:HepBPCRstep4.2.JPG|500px|center|thumb|<p align="justify"> | + | [[File:HepBPCRstep4.2.JPG|500px|center|thumb|<p align="justify">Figure 1: A smear was the result. In the blue circle a possible positive result can be seen.</p>]] |
The words with the figure are carefully chosen, because similar bands should show in lanes 2-4, in which they are absent. Therefore, we need an alternative approach in which first the annealing is done after which the primers are added for elongation. | The words with the figure are carefully chosen, because similar bands should show in lanes 2-4, in which they are absent. Therefore, we need an alternative approach in which first the annealing is done after which the primers are added for elongation. | ||
| Line 68: | Line 94: | ||
• Final attempt PCR step 4 | • Final attempt PCR step 4 | ||
| - | The final attempt was done using gel purified | + | The final attempt was done using gel purified PCR products. This did not yield a better result than before. Therefore, a whole new strategy needs to be created. |
| Line 108: | Line 134: | ||
''14.August'' | ''14.August'' | ||
| - | * the PCR product of the 2nd PCR reaction was loaded on a big gel and the bright band was | + | * the PCR product of the 2nd PCR reaction was loaded on a big gel and the bright band was separated from second band present in the gel. those bands where cut out seperately and purified |
* the PCR product (PCR purified as well as the cut out fragments) where ligated into Bba_J04500 and transformed with DH5α | * the PCR product (PCR purified as well as the cut out fragments) where ligated into Bba_J04500 and transformed with DH5α | ||
-> no growth of the transformants | -> no growth of the transformants | ||
| Line 114: | Line 140: | ||
''15.August'' | ''15.August'' | ||
| - | * the two | + | * the two separate samples (the two bands) from the gel extraction where checked with the primers of the 2nd PCR reaction |
-> only one sample reacts on the primers | -> only one sample reacts on the primers | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | '''CCMV Stability experiments''' | ||
| + | |||
| + | |||
| + | ''11th - 14th August'' (Mark) | ||
| + | <ul> | ||
| + | <li>Ultracentrifuge 4 times</li> | ||
| + | <li>Followed protocol of purification CCMV</li> | ||
| + | <li>Total end volume is 15 ml</li> | ||
| + | </ul> | ||
| + | |||
| + | |||
| + | ''14th – 15th August'' (Mark) | ||
| + | <ul> | ||
| + | <li>FPLC with the S400 column</li> | ||
| + | <li>3 runs with 4 ml sample</li> | ||
| + | <li>All good separation with them with good concentration</li> | ||
| + | </ul> | ||
---- | ---- | ||
[[https://2012.igem.org/Team:Wageningen_UR/Journal/week15 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week17 next week]] | [[https://2012.igem.org/Team:Wageningen_UR/Journal/week15 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week17 next week]] | ||
Latest revision as of 20:19, 25 September 2012
week 16: 13 august - 19 august
Office work
Lab work
New competent cell used for protein expression (JM-109) was made this week, transformation efficiency was checked, which was 1*10^6.
CCMV
13th August (Hugo)
Colony PCR on IPTG_CCMV_NEG transformant colonies and 5 red BBA_J3901 colonies.
Grew two colonies of IPTG_CCMV_NEG overnight for miniprepping.
14th August (Hugo)
Robert miniprepped the overnight cultures that were inoculated at 13-08. Then I ran a digestion check with EcoRI + PstI.
Sent sample with CCMV neg in the chloramphenicol backbone for sequencing (GATC).
Prepared iron-free E. coli medium.
16 & 17th August (Hugo)
Prepared JM 109 competent cells.
TuYV
- Last week's 'brick' minipreps were PCR checked for the correct insert. Results were unclear at first, but a second check gave positive results for the TuYV Coat Protein brick (1), the CP + Histidine tag brick(1H), and the CP + readthrough brick (4)! Only the CP + readthrough + His-tag(4H) brick wasn't found.
- The second trial of putting TuYV constructs after IPTG induced promoter turned out to be a failure.
Hepatitis B
- 13 August (Lisa, Kees)
• SDS PAGE and VLP production
The cells (pre-induction and induced) were lysed using French press and analysed using SDS PAGE. A clear induction was shown, but because of a poor ladder, the HepB subunits could not be pointed out.
VLP production is continued according to the Hepatitis B VLP formation protocol [link]. We designed this protocol based on literature and the CCMV protocol.
- 16 August (Lisa, Kees)
• Finishing VLP formation
The dialysis was finished yesterday. Today, the dialysed solution was ultra-centrifuged, after which we dissolved the pellet in Formation Buffer, as little as we could use, about 0.2mL per centrifugation tube. An aliquot of all the tubes was made and stored at 4˚C until EM conformation.
since colony PCR of the HepB core protein in the Bba_J04500 backbone showed no results, the samples where minipreped to check it again
Hepatitis B outside modification
- 13-14 August (Kees)
• Step 4 with alternative PCR program
Step 1-3 were repeated on 13 and 14 August. The DNA was gel purified after each step, enhancing the wanted product, though not excluding wrong products. Step 4 was repeated with an annealing step before adding the primers. Protocol: 1: 98˚C; 2min 2: 98˚C; 30s 3: 67˚C; 30s 4: 72˚C; 30s 5× from step 2 Added primers while: 5: 72˚C; 10min 6: 98˚C; 30s 7: 60˚C; 30s 8: 72˚C; 40s 35× from step 6 9: 72˚C; 10min 10: 4˚C; ∞
The words with the figure are carefully chosen, because similar bands should show in lanes 2-4, in which they are absent. Therefore, we need an alternative approach in which first the annealing is done after which the primers are added for elongation. Apparently, this approach does not work. The two fragments do not anneal properly, maybe because the Tm from the whole overhang is too high, 68-74˚C depending on calculation method used. Or, the chance of this whole event occurring as planned is just too low.
- 16 August (Kees)
• Final attempt PCR step 4
The final attempt was done using gel purified PCR products. This did not yield a better result than before. Therefore, a whole new strategy needs to be created.
Hepatitis B inside modification
13.August
- 2nd step of the PCR reaction
-> the gel shows the slightly bigger fragment of the second step compared to that of the 1st step at the expected size. A smear is visible in the lanes containing the samples of the 2nd step.
14.August
- 3rd step of the PCR reaction
-> a fragment was present of about the right size - but no conclusion was possible because of a strong band in the negative control of about the same size
15.August
- PCR purification of the 1st step of the PCR reaction and again 2nd step of the PCR reaction using this sample as a template
-> fragments resulting from this reaction don't have the right size
- 4th step of the PCR reaction
-> this step did not work
16.August
- repeat step 4
- check of the activity of sample step 3 upon primer step 3
-> there was no band visible on the gel - no conclusion possible
GFP modification
13.August
- 2nd (final) step of the PCR reaction
-> the gel shows slightly bigger fragments of the expected size - but also a very weak second band and a smear in these samples
14.August
- the PCR product of the 2nd PCR reaction was loaded on a big gel and the bright band was separated from second band present in the gel. those bands where cut out seperately and purified
- the PCR product (PCR purified as well as the cut out fragments) where ligated into Bba_J04500 and transformed with DH5α
-> no growth of the transformants
15.August
- the two separate samples (the two bands) from the gel extraction where checked with the primers of the 2nd PCR reaction
-> only one sample reacts on the primers
CCMV Stability experiments
11th - 14th August (Mark)
- Ultracentrifuge 4 times
- Followed protocol of purification CCMV
- Total end volume is 15 ml
14th – 15th August (Mark)
- FPLC with the S400 column
- 3 runs with 4 ml sample
- All good separation with them with good concentration
 "
"