Team:Evry/GB
From 2012.igem.org
| (90 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<html> | <html> | ||
| - | <h1><b>The | + | <h1 align=center><b>The GoldenBricks:</b> A fast cloning technique <br> for the PartsRegistry</h1> |
| - | < | + | <center><img src="https://static.igem.org/mediawiki/2012/9/9e/S%C3%A9lection_163.png" width=250px></center> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<h1>Introduction</h1> | <h1>Introduction</h1> | ||
| Line 15: | Line 11: | ||
<h2>Context</h2> | <h2>Context</h2> | ||
| - | |||
| - | <p> | + | <p>Cloning is probably the most tedious and less rewarding task when engineering living entities through synthetic biology. With the development of the restriction enzymes and efficient DNA ligases, assembling small and large DNA pieces has become a common practice in biology, but as everyone is using their own sets of enzyme and assembly method, making large DNA constructs is still a challenge today. The first indisputable advance in the domain came with the invention of the first standardized DNA cloning method: the BioBricks [1]. The standard BioBrick format (BBF RFC 10) make possible to clone together all kind of genetic parts (called "bricks") using only four different enzymes: EcoRI, XbaI, SpeI and PstI. But the real power of BioBrick standardization lays in the possibility to build database made of these DNA pieces that anyone can be assembled through a standard method. The PartsRegistry today contains almost over 3,000 basic characterized biological parts [2] and is a sources of inspiration for thousands of biologists around the world.<p> |
| - | <p> | + | <p>If biobricks have opened new perspectives for engineering organisms and have proven their efficiency over a decade, their use is limited by the fact that the fragments can only be assembled one by one. On the top of this, the process is very difficult to automatize, because of the number of DNA purification step required. IGEMers today spend most of their time assembling genetic parts together, leaving little time for characterizing them and testing their systems.</p> |
| + | <p>Several cloning techniques capable of assembling multiple fragments at the time have been invented since the rise of the Biobrick format. The most popular one is probably the one known as the Gibson method [3] that can be used to assemble up to four fragments at the time on a regular basis. The Golden Gate technique [4] (>20 fragments at the time) and its new evolution, the MoClo [5] (47 fragments in two times reported) are leading the way of another kind of cloning technique based on type II restriction enzymes. More efficient than the Gibson cloning, the Golden Gate also have the huge avantage that it can assemble more parts at the time, no matter their size. Moreover, Golden Gate do not imply any DNA modifications, which noticeably reduce the probability of having a mutation at the ligation scar.</p> | ||
| - | < | + | <p>If these new techniques dramatically speed-up the assembly of DNA pieces, to our knowledge, no reported work has been carried on standardizing these methodologies. So far, biologists using Golden Gate engineer new overhangs and create a new library each time they are making a different construct.</p> |
| - | <p>The | + | |
| + | <h2>From Golden Gate and BioBricks to GoldenBricks</h2> | ||
| + | |||
| + | <p>The BioBricks are a collection of parts that can be assembled one by one in a generic way. Golden Gate is a technique that enable the assembly of several tens of parts simultaneously. Taking inpiration from both technique, the 2012 Evry iGEM Team invented and developped a new methodology called GoldenBricks, merging the power of the two techniques while keeping the compatibility toward the old RFC 10 BioBrick format and preserving the possiblilty to make standard GoldenGate assembly simultaneously with GoldenBrick assembly, using the same vectors.<p> | ||
| + | |||
| + | <p>GoldenBricks technique is a one-shot cassette construction using DNA parts coming either from a plasmid distribution or from PCR product, engaging 5, 7 and possibly more different parts. Moreover, GoldenBricks works for both eukaryotes and procaryotes DNA construction with the very same protocol. If a non classical assembly is required (as for testing the strength of a terminator) a new "split construction" method based on standard plasmids make possible to assemble parts in a non classical way.</p> | ||
<h2>Perspectives</h2> | <h2>Perspectives</h2> | ||
| - | <p>Such technique offers | + | <p>Such technique offers great perspectives for the future of iGEM and of the PartsRegistry in general. First, it would make possible the fast and simple assembly of complete cassettes using less expensive equipment, less toxic chemical and less sequencing runs than the BioBrick assembly. Second, it would make fast and efficient cloning accessible for both researchers as well as for less experimented users, such as high school iGEMers or biohackers thanks to the simplicity of its protocol. Third, the creation of large expression cassettes using this method is cheaper and faster than synthesizing the construct at the present day, which would guaranty the interest for a DNA database compared to <i>de novo</i> synthesis for the years to come. And last, this method is a lot more easy to automatize unlike the RFC 10 biobrick format.</p> |
| - | <h1> | + | <h1>Theory</h1> |
| - | <h2>Requirements for the development of the new standard</h2> | + | <a name="requirement" ></a><h2>Requirements for the development of the new standard</h2> |
| - | <p>In this work, | + | <p>In the continuation of what we wrote previously and in order to stay in the progression of the methodology we have tried in this work to stay as close as possible to the biobrick format, to keep GoldenBrick parts compatible with the RFC 10 standard assembly, while opening new perspectives to increase the speed and the easiness of cloning. These constraints imposed several requirements for this new standard.</p> |
| + | |||
| + | <p>First, the new assembly method should:</p> | ||
<ol> | <ol> | ||
| - | <li> | + | <li>Keep the compatibility with the current BBF RFC 10 standard</li> |
| - | <li> | + | <li>Be compatible with a database approach</li> |
| - | <li>Use the standard registry | + | <li>Minimize the number of different prefixes and suffixes</li> |
| + | <li>Use the standard registry plasmids and negative cloning control</li> | ||
</ol> | </ol> | ||
| - | <p> | + | <p>To improve the cloning speed, we would also like to:</p> |
| - | <ol start= | + | <ol start=5> |
| - | <li> | + | <li>Make one step Golden Gate cassette cloning possible</li> |
| - | + | <li>Allow to check the construct with a single enzyme digestion</li> | |
| - | <li>Allow to check the construct with a single digestion</li> | + | <li>Improve the compatibility with Gibson by removing the NotI site</li> |
| - | <li>Improve the compatibility with Gibson</li> | + | <li>Provide a solution for non standard assemblies</li> |
| - | <li>Provide a solution for non standard | + | |
</ol> | </ol> | ||
| - | <p> | + | <p>As we will demonstrate, the GoldenBrick format we have established meet all these requirements. To understand on what basis we have created it, we are now going to explain in this section the principles that led the design of this format before we analyze the sequences of the GoldenBrick prefixes and suffixes and finally discuss what can be done with this technique. |
| - | <h2> | + | <h2>A brief introduction on type II restriction enzymes</h2> |
| - | < | + | <p>Golden Gate cloning, (also known as type II restriction enzyme cloning) rely on a specific familly of enzyme (known as the type IIS enzymes) very different from the ones we are used to. The characteristic of type IIS enzymes from the cloning point of view is that these enzymes cut the DNA outside their recognition locus, no matter the sequence of the place where they cut. A very small subtype of them can be used for cloning purpose, since they generate overhangs on one side of the recognition site only. The ones used for Golden Gate and MoClo cloning are BsaI and BbsI.</p> |
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:202px;"><a href="https://static.igem.org/mediawiki/2012/0/0b/S%C3%A9lection_164.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/0/0b/S%C3%A9lection_164.png" width="200" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/0/0b/S%C3%A9lection_164.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 2: The recognition pattern of BsaI and cutting site.</div></div></div></div> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <p>The fact that these enzymes cut outside its recognition site gives them two significant advantages:</p> | ||
| + | <ul> | ||
| + | <li>You can generate as many different overhang you want with a single enzyme within the same tube</li> | ||
| + | <li>You can engineer your fragment so that it cannot be cutted again after it has ligated.</li> | ||
| + | </ul> | ||
| + | |||
| + | <p>Thanks to this last point, it is possible to cut and ligate the DNA in the mean time within the same tube (BsaI works in the T4 ligase buffer), and do not require any DNA purification. For this reason, the ligation is achieved with a better yield and with less mutation than with traditional type II enzymes cloning methods.</p> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:502px;"><a href="https://static.igem.org/mediawiki/2012/9/90/S%C3%A9lection_165.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/9/90/S%C3%A9lection_165.png" width="500" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/9/90/S%C3%A9lection_165.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure 3: Principle of the Golden Gate cloning method.</div></div></div></div> | ||
| + | |||
| + | <p>Since the small BsaI fragment cut can re-ligate with the mother strand, the assembly is a lot more efficient if the tube is alternatively placed at 37°C, when the BsaI cuts and then placed at 16°C for a ligation time, using a thermocycler. This technique drive the equilibrium towards the product formation.</p> | ||
| + | |||
| + | <h2>Analysis of a classical synthetic biology construction</h2> | ||
| + | |||
| + | <p>A traditional construct in synthetic biology is made of the repetition of the following elements:</p> | ||
| + | |||
| + | |||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:402px;"><a href="https://static.igem.org/mediawiki/2012/6/6e/Constructs.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/6/6e/Constructs.png" width="400" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/6/6e/Constructs.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 4: Schematic of the different elements assembly in a standard synthetic biology device.</div></div></div></div> | ||
| + | |||
| + | <p>At the moment, GoldenBrick only allows the assembly of a single cassette at the time. In order to create the full system, the assembly of the different cassettes together have to be conducted afterwards with the biobrick assembly, Gibson assembly - that is very efficient to assemble long fragments - or ligation independant cloning [6]. Focusing closer on a single cassette, we can notice that we have four different kinds of junctions:</p> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:502px;"><a href="https://static.igem.org/mediawiki/2012/2/26/Constructs_anotated.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/2/26/Constructs_anotated.png" width="500" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/2/26/Constructs_anotated.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 5: In a single cassette, we can notice four different kinds of junctions. If we want the repetition of the {RBS-CDS} unit to be possible, the prefix of the RBS have to be compatible with the suffix of the gene.</div></div></div></div> | ||
| + | |||
| + | <p>As we can notice on this picture, there is four different kinds of junctions in an expression cassette. In Golden Gate, the junctions are DNA overhangs that can be freely engineered thanks to the capacity of Type IIS enzymes to cut outside their recognition site, no matter what sequence is there. Therefore, we should engineer four different overhangs. The first overhang is dedicated to the ligation of the plasmid suffix with the promoter prefix (OH1). Similarly, the 5th one is dedicated to ligate the terminator suffix with the backbone prefix (OH5). In a bacterial device, the scar between the RBS and the gene (OH3) has to be precisely controlled because the distance between the two elements is critical for correct protein expression. This scar will remain the same than in the biobrick assembly, but it will be generated by a type IIS enzyme as for the other overhangs. Finally, the overhang that link the RBS prefix to the promoter suffix (OH2) and the protein suffix to the terminator prefix (OH4) have to be compatible to make the repetition of several {RBS-Protein} units in a single cassette (OH2=OH4) possible. We will see later how we can achieve control of this repetition.</p> | ||
| + | |||
| + | <p>To conclude, we have five different kinds of parts, which implies to create five different prefixes and five different suffixes with four different overhangs, which will help to assemble the five different DNA parts in the correct order.</p> | ||
| + | |||
| + | <p>In addition, we would like to generate a scar that can be digested by another enzyme, in order to control whether all the elements we intended to insert are present in the cassette or not, before sequencing it. As we will see later this element is critical for the selection of the clone that have inserted all the genes we want and avoid sequencing incorrect constructions. In order to generate such a scar, the overhangs 1, 2, 4 and 5 has been engineered in the middle of a BbsI site. When two pieces ligates, they recreate a BbsI restriction site, and the construct can be checked with a simple BbsI digestion.</p> | ||
| + | |||
| + | <p>One last condition to fulfill to get a RFC 10 compatible brick after GoldenBrick cloning is to leave no illegal restriction site inside the brick after the assembly of the cassette. However, we should keep the as similar as possible the BioBrick extensions around each elements leaving the four usual enzyme restriction sites, so that the brick can be still assembled using RFC 10 BioBrick method. The only drawback is that if several GoldenBrick parts are assembled with RFC 10 standard, they cannot be assembled with other GoldenBrick parts afterwards, because the BioBrick assembly will leave illegal BsaI site inside the casette.</p> | ||
<h2>The proposed new set of Golden Bricks extension</h2> | <h2>The proposed new set of Golden Bricks extension</h2> | ||
| - | < | + | <p>Having all these design principles in mind and meeting the requirements we have proposed, we designed the new GoldenBrick prefix and suffix with the following sequences:</p> |
| - | |||
| - | < | + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:635px;"><a href="/File:GoldenBricks.png" class="image"><img alt="" src="/wiki/images/e/ed/GoldenBricks.png" width="633" height="286" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:GoldenBricks.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure 6: Sequence and design of the different prefixes and suffixes used in the GoldenGate cloning method. The presence of the EcoRI, XbaI, SpeI and PstI keep the compatibility with Biobrick cloning while the BsaI site enable single shot GoldenGate assembly. The BbsI sites serve as a single digestion control.</div></div></div></div> |
| - | < | + | <p>This design fulfills all the requirements enumerated before and ensure that the different elements will be assembled in the correct order. We also generated a scar that can be digested afterwards with the BbsI enzyme, so that the correct insertion of the different elements can be checked afterwards by doing a single digestion. This also enable the polymerization of the {RBS-CDS} units, as we will discuss in the next paragraph.</p> |
| - | <h2> | + | <h2>Control over the polymerization</h2> |
| + | |||
| + | <p>The only difference with a standard GoldenBrick protocol is the capacity for the RBS-protein cassette to polymerize. This polymerization capacity makes the power of the technique because it enables to make polycistronic mRNAs no matter what gene is inserted. However, a control mechanism is required in order to create polycistrons or when we want a single gene to be inserted in the cassette.</p> | ||
| + | |||
| + | <p>In order to achieve such a control, several parameters in the protocol can be ajusted to move the equilibrium from monocistrons to polycistrons. The first and the most obvious one is the stochiometry of the different elements. If the promoters and terminators are in large excess compared to the RBSs and the gene, the balance will be in favour of monocistrons. On the contrary, if the amount or RBSs and coding sequence in dominant, the cassette will tend to contain two or more genes inserted inside.</p> | ||
| + | |||
| + | <p>On the other hand, the number of cycle and the ligation time will also influence the polymerization. Polycistrons are not likely to appear when the number of cycle is small and the ligation time short, but they will become more and more important as we increase these parameters.</p> | ||
| + | |||
| + | <p>Most of the work carried on by our team is now focused on optimizing the protocol. We will release soon a set of standard protocols for mono and polycistrons optimized to reduce the heterogeneity of the polymerization length.</p> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:502px;"><a href="/File:GoldenBrick-polymerization.png" class="image"><img alt="" src="/wiki/images/1/10/GoldenBrick-polymerization.png" width="500" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:GoldenBrick-polymerization.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 8: a. The vector, promoter, and terminator are in excess: the assembly produces mostly monocistrons. b. The protein sequences and RBS are in excess: the assembly produces mostly polycistrons.</div></div></div></div> | ||
| + | |||
| + | |||
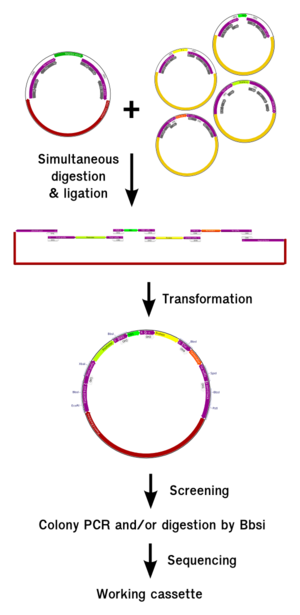
| + | <h2>Assembly procedure</h2> | ||
| + | |||
| + | <div class="thumb tright"><div class="thumbinner" style="width:302px;"><a href="/File:GOldenbrick_global_thumb.png" class="image"><img alt="" src="/wiki/images/thumb/f/f3/GOldenbrick_global_thumb.png/300px-GOldenbrick_global_thumb.png" width="300" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:GOldenbrick_global_thumb.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 9: Summary of the GoldenBrick procedure</div></div></div> | ||
| + | |||
| + | |||
| + | <p>Assembling parts using the GoldenBrick method is a lot easier than for BioBricks and also five to ten times faster. We are now going to explain the procedure from the theoretical point of view. For the complete protocol, you should refer to the material and methods section.</p> | ||
| + | |||
| + | <p>Because it requires a very small amount of DNA, the plasmid source can be taken directely from a DNA distribution suck as the PartsRegistry distribution, without requiring the need of a DNA preparation before. Once the plasmids are resuspended, the experimentalist mix them together in a mix containing the BsaI enzyme and T4 ligase and put them in the thermocycler for 5 hours. At the end, the product can be directly transformed and plated. A first round of screening on the good polymerization length can be made using colony PCR with the standard VF2 and VR primers. If required, a second step of screening can be made after miniprepping using BbsI. Given the very small rate of mutation, only two clones with the good digestion or colony PCR signature can be sent for sequencing; they will contain the correct construct with a very high probability.</p> | ||
| + | |||
| + | <p>This technique however requires to screen an important number of clones because we don't have a total control over what is assembled inside the cassette, because of the freedom we left for the RBS-Protein entities to polymerize. We will show in the next paragraph how we can gain a relative control over it. To reduce the costs of screening, we encourage the use of colony PCR over mini-preping and digestion when possible.</p> | ||
| + | |||
| + | |||
| + | <h1>Other advantages of GoldenBricks over any other standard cloning techniques</h1> | ||
| + | |||
| + | <h2>Non standard assembly and "split-construct" vectors</h2> | ||
| + | |||
| + | <p>If you are willing to create a non standard assembly or to control very precisely RBS-protein pairing over a polycistrons, you might like to create the construct in two times and assemble them together in a second step. This is why we have created the "split-construct" vectors. The split-contruct vectors are similar to the goldenbrick vectors except that they contain the OH2-OH5 or the OH1-OH4 overhangs instead of the OH1-OH5 overhangs as the GoldenBrick plasmid.</p> | ||
| + | |||
| + | <p>These vectors enable the assembly of shorter casette containing only a promoter, and a serie of RBS-protein for the "split-construct" 1 vector, or RBS-protein terminator for the "split-construct" 2 vector. This way, if a non standard biobrick assembly has to be carried out in between the two pieces or if a long polycistron has to be created, they can be assembled using the standard EcoRI, XbaI, SpeI, PstI cloning or Gibson assembly, but several cloning step will be saved by using GoldenBricks compared to the standard assembly of the full cassette.</p> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:802px;"><a href="https://static.igem.org/mediawiki/2012/1/19/Split-constructs.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/1/19/Split-constructs.png" width="800" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/1/19/Split-constructs.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 10: Description of the construction of a non classical cassette for creating a device that test the strength of a terminator using split-vector GoldenBricked mixed with classical or Gibson assembly. The GFP and RFP ratios are measured to evaluate the strength of a terminator the classical way. Two cloning steps are avoided using this procedure.</div></div></div></div> | ||
| + | |||
| + | |||
| + | <h2>GoldenBrick make standard part DNA shuffling possible</h2> | ||
| + | |||
| + | <p>Aside from the dramatic shortening of the cloning time, the decrease of the costs and the easiness of GoldenBricks over Biobricks, one of the most noticeable improvement is undoubtedly the possibility to make DNA shuffling using a standard format. Let us take the example of a complex system such as a toggle-switch, in which you would like to optimize the expression of the protein repression of its two components to place the system in the working area of the phase domain.</p> | ||
| + | |||
| + | <p>Using the Biobrick format, you will probably have to make tens of different constructs with different RBS to find a condition in which the toggle-switch will work. Using GoldenBricks, you can make all of them in a single tube, in a single step. Moreover, you may eventually find a way to screen them directly after the cloning avoiding the need of purifying and sequence all the clones that couldn't work.</p> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:802px;"><a href="https://static.igem.org/mediawiki/2012/3/31/DNA-shuffeling.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/3/31/DNA-shuffeling.png" width="800" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/3/31/DNA-shuffeling.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 11: Description of a DNA shuffling experiment in order to optimize the expression of a single gene. 4 Promoters and RBSs are mixed together in a single tube and give rise to 16 construct in a single tube in a single step. The clone expression the enzyme at the correct level can be screened directly on the transformation plate.</div></div></div></div> | ||
| + | |||
| + | <p>Many other use of DNA shuffling can be found in the literature especially for gene expression optimization and screening for working enzymes. We expect that DNA shuffling over standard parts is going to find many applications in the future of iGEM projects.</p> | ||
| + | |||
| + | <h2>GoldenBrick is still compatible with Biobrick format</h2> | ||
| + | |||
| + | <p>If the situation requires the part to be assembled with the standard RFC 10 format, the new GoldenBrick parts can be assembled the very same way than BioBricks. The only limitation is that BioBrick assembly of GoldenBrick parts leave some scars containing the BsaI restriction site inside, and the produced parts cannot be reassembled with other parts using Golden Gate. No simple way of fixing this issue has been found.</p> | ||
| + | |||
| + | <h2>GoldenBricked plasmids are compatible with standard Golden Gate assembly</h2> | ||
| + | |||
| + | <p>The GoldenBricked plasmids can be used to assemble contructions using classical Golden Gate. This option has been tested in our team and it works fine. See the result section for more results on the use of GoldenBrick vectors with the construction of the IaaH-Pep2A-IaaM fusion protein.</p> | ||
| + | |||
| + | <h2>GoldenBrick also assemble PCR products and shorten the assembly time</h2> | ||
| + | |||
| + | <p>Someone that want to create a construct not having the time or the need of cloning his gene into a vector first can still assemble them using GoldenBrick. Although this practice wouldn't be encouraged for standards parts, it can become really useful when trying libraries of different genes in order to find a working one. GoldenBricked parts in plasmid and PCR GoldenBricked parts can be mixed together and assembled the very same way. The user will just have to take care to submit his working parts afterwards. We recommend to amplify new parts flanked by its GoldenBricks prefix and suffix, mix them with the GoldenGate assembly mix in one hand, and digest them in EcoRI and PstI on the other hand and ligate them in pSB1C3 for submitting the GoldenBricked part to the registry. | ||
| + | |||
| + | <h2>Classical GoldenGate can be carried out in the mean time than GoldenBrick assembly to create fusion proteins within the construct</h2> | ||
| + | |||
| + | <p>GoldenGate in general and GoldenBricks in particular are very efficient to assemble fusion proteins. This is because one can design every overhang it wants and therefore fuse protein domains without leaving any scar. New fusion proteins can be created in the meantime they are assembled in the cassette, using an aditional scar designed by the experimentalist. We also recommand to submit the new fusion protein part separately to contribute to the registry.</p> | ||
| + | |||
| + | <h2>Finally, GoldenBricks works for eukaryotes chassis as well as with prokaryotes</h2> | ||
| + | |||
| + | <p>As we have seen all along this article, this design of GoldenBricks have been designed for procaryotes, but it can work very well for eukaryotes. In eukaryotes, promoters can include or not a 5'UTR in their sequence to enhance the protein expression. In the case there is no 5'UTR, we can use the same prefix and suffix than the prokaryote promoters and include the 5'UTR with the prokaryotes prefix and suffix of a prokaryotic RBS.</p> | ||
| + | |||
| + | <p>In order to make polycistrons, we can use an Intermediate Ribosome Entry Site (IRES), cloned between an prokaryote RBS prefix and suffix.</p> | ||
| + | |||
| + | <p>Finally, the polyA and the polyA capping cassette with a polymerase terminator can be flanked by the prokaryotes terminators sites. In the case one don't want to include a polyA capping, he can use the "split construct" 1 or in the case there is not terminator, the one on the plasmid will take care of stopping the RNA polymerase extension.</p> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:802px;"><a href="https://static.igem.org/mediawiki/2012/0/0a/GoldenBricks-eukaryote.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/0/0a/GoldenBricks-eukaryote.png" width="800" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/0/0a/GoldenBricks-eukaryote.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 12: GoldenBricks prefixes and suffixes for eukaryotic constructions.</div></div></div></div> | ||
| + | |||
| + | <h1>Results</h1> | ||
| + | |||
| + | <h2>Construction of the library</h2> | ||
| + | |||
| + | <p>In order to test the GoldenBrick technique efficiently, and for easiness of screening and statistics, we have developed a set of GoldenBricked parts (see Material & Methods) with fluorescent proteins markers. We have created and submitted the following parts:</p> | ||
| + | |||
| + | <ul> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812050">K812050:</a> A GoldenBricked version of pSB1C3 with J04450 as negative cloning control (status=sent)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812051">K812051:</a> A GoldenBricked version of pSB1K3 with J04450 as negative cloning control (status=constructed)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812053">K812053:</a> A GoldenBricked version of the strong RBS B0034 (status=sent)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812054">K812054:</a> A GoldenBricked version of the RFP E1010 (status=sent)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812055">K812055:</a> A GoldenBricked version of the terminator B0015 (status=sent)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812056">K812056:</a> A GoldenBricked version of the pLac R0010 promoter (status=sent)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812057">K812057:</a> A GoldenBricked of an sfGFP protein (status=constructed)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812058">K812058:</a> A GoldenBricked of medium strenght RBS J61107 (status=constructed)</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812058">K812059:</a> A GoldenBricked of week RBS J61117 (status=constructed)</li> | ||
| + | </ul> | ||
| + | |||
| + | <p>All the parts written here have been successfully constructed and sequenced and the ones marked as "sent" has been sent to the PartsRegistry.</p> | ||
| + | |||
| + | <a name="GB" ></a><h2>Creation of a simple cassette</h2> | ||
| + | |||
| + | <p>We have started experimenting on the GoldenBrick assembly method. For the moment, the results are not optimal so we are not publishing them yet. Better results are comming soon. For the moment, we have about 1% of correct clones.</p> | ||
| + | |||
| + | <a name="GG" /></a><h2>Working with regular GoldenGate using GoldenBricks vectors</h2> | ||
| + | |||
| + | <p>In order to assemble the fusion proteins we need for our project, we carried out two assemblies of fusion proteins using Golden Gate in our Golden Gated vectors. This paragraph reports the construction of the IaaH-Pep2A-IaaM fusion protein in the GoldenBrick plasmid K812050 using standard Golden Gate protocol. This protocol hadn't been optimized at the moment. However, out of 16 cloned on which we did colony PCR, 10 of them had the good size. We sequenced 4 of them and obtained the sequence expected.</p> | ||
| + | |||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:702px;"><a href="https://static.igem.org/mediawiki/2012/thumb/4/40/IaaH-Pep2A-IaaM.png/800px-IaaH-Pep2A-IaaM.png" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/thumb/4/40/IaaH-Pep2A-IaaM.png/800px-IaaH-Pep2A-IaaM.png" width="700" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="https://static.igem.org/mediawiki/2012/thumb/4/40/IaaH-Pep2A-IaaM.png/800px-IaaH-Pep2A-IaaM.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 13: Colony PCR of 15 clones from the transformation plate. The ones marked with a star were sequenced and proved to be correct. The one with an exclamation mark was sequenced and had only one of the CDS inserted.</div></div></div></div> | ||
| + | |||
| + | <p>This experiment demonstrates the possibility of doing GoldenGate in the GoldenBricks plasmids K812050 (and similarly in K812051) in a very efficient way.</p> | ||
| - | |||
<h1>Material and methods</h1> | <h1>Material and methods</h1> | ||
<h2>Construction of the library</h2> | <h2>Construction of the library</h2> | ||
| + | |||
| + | <p>The GoldenBricked parts were constructed amplifying the corresponding biobricks using primers flanked by the appropriate GoldenBricks extensions, using Phusion (Thermo Scientific) or Q5 (NEB) DNA polymerase in a 30 cycle reaction. The PCR products were PCR purified and then digested using FastDigest® EcoRI and PstI (Thermo Scientific). The standard plasmid pSB1C3 (or pSB1K3) were digested similarely. Both vectors and inserts were PCR purified and then ligated using T4 DNA ligase (Thermo Scientific) before being transformed with home made competent cells (chemical or electroporation). Four white clones were inoculated and sequenced.<p> | ||
| + | |||
| + | <p>The either red or non red colonies were minipreped and then digested with BsaI for screening. All the clones that had the correct digestion pattern were sequenced and gave the correct sequence.</p> | ||
| + | |||
| + | <p>The RBSs were synthetized in the form of two matching oligos. A mixture of 10 µM of the two oligo was incubated at 98°C for 2 min and then cooled down to 4°C with a ramp of -1°C/s. They were then PCR purified, digested and PCR purified again before beeing ligated in pSB1C3.</p> | ||
<h2>Assembly protocol</h2> | <h2>Assembly protocol</h2> | ||
| - | <h1> | + | <p>The protocol given here hasn't been optimized yet. We are only starting the development of this new technology. It is given here as a matter of indication. Some optimized protocol will be available soon.</p> |
| + | |||
| + | <p>About 75 ng of each plasmid (more accurate ratios will be optimized) were mixed together with 2.5 units of BsaI enzyme (NEB) and 15 units of T4 DNA ligase (Thermo Scientific) in a total volume of 15 µL of standard T4 ligation buffer. The tubes were then placed in the thermocycler with the following steps:</p> | ||
| + | |||
| + | <ol> | ||
| + | <li>37°C, 2 min (digestion)</li> | ||
| + | <li>16°C, 5 min (ligation)</li> | ||
| + | <li>Goto 1, 50 times</li> | ||
| + | <li><ul><li>Option 1: if the construct does not contain any BsaI site: 50°C 5 min (reduce the background with a final digestion)</li> | ||
| + | <li>Option 2: if the construct contains BsaI site: 16°C 2h (reconstitute the BsaI site)</li></ul></li> | ||
| + | <li>80°C, 5 min (heat inactivation)</li> | ||
| + | </ol> | ||
| + | |||
| + | <p>5 µL of the product was transformed with 50 µL of home made chemically competent cells and recovered for 1 hour. The cells were concentrated and then plated.</p> | ||
| + | |||
| + | <h1>Perspectives for GoldenBricks and the PartsRegistry</h1> | ||
| + | |||
| + | <p>When the complete protocols will be established, our team will submit a BBF RFC request for the registry. If the RFC get accepted and if the tenant of the PartsRegistry thinks this is a valuable format, we can think of migrating the registry to this new format. This can be done in two complementary ways: the first one is to ask every user to submit their new part in the new GoldeBrick format, and the second is to take advantage of the huge community working on the PartsRegistry and especially the iGEM competition. In order to migrate the already existing parts faster, we can add in the requirements for the gold medal to resubmit already exsiting parts in the new format. With an average of 200 teams in the years to come and asking each of them to resubmit two goldenbricked parts, we can cover the 2000 most useful parts within two or three years, because the teams will GoldenBrick the parts they are interested in for their assembly, so they will probably always re-submit more than two.</p> | ||
| + | |||
| + | <p>We believe that implementing such a method in the partsregistry will speed up the assembly process and give a new shine to this database that severely suffers from the gene synthesis and PCR based methods competition. This will leave more time for researchers and iGEMers for characterizing the parts, and the partregistry will also benefits from an increase of parts characterization, which is very beneficial to the community. Overhall, the idea of standardized parts that tends to loose its strength with the time will gain again in interest in the long run.</p> | ||
<h1>Conclusion</h1> | <h1>Conclusion</h1> | ||
| + | |||
| + | <p>This year our team created a new cloning format for the PartsRegistry that dramatically speeds up the cloning time and opens new perspective. This includes simple protein fusion methods and DNA shuffling that were not possible using the current BioBrick format. At the moment, we are working very hard to demonstrate the possibilities of the technique and come up with efficient and reliable protocols. If the process proves its interest, we will submit an RFC to the PartsRegistry and propose the GoldenBrick progressively substitute to the current BioBrick format, which will renew the interest for people on standardized biological parts and standardized assembly over gene synthesis.</p> | ||
| + | |||
| Line 84: | Line 254: | ||
<p id="references">References:</p> | <p id="references">References:</p> | ||
<ol> | <ol> | ||
| - | <li><i> | + | <li><i>Knight TF</i>, Idempotent Vector Design for Standard Assembly of BioBricks. 2003, Tech rep, MIT Synthetic Biology Working Group Technical Reports.</li> |
| + | <li>Source: http://partsregistry.org/cgi/partsdb/Statistics.cgi</li> | ||
| + | <li><i>Gibson D</i>, One-step enzymatic assembly of DNA molecules up to several hundred kilobases in size. 2009, Nature Publishing Group: doi:10.1038/nprot.2009.77</li> | ||
| + | <li><i>Engler C, Gruetzner R, Kandzia R, Marillonnet S</i>, Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. 2009, PLoS ONE 4(5): e5553. doi:10.1371/journal.pone.0005553</li> | ||
| + | <li><i>Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S</i>, A Modular Cloning System for Standardized Assembly of Multigene Constructs, 2011, PLoS ONE 6(2): e16765. doi:10.1371/journal.pone.0016765</li> | ||
| + | <li><i>Charalampos A & J. de Joung P</i>, Ligation-independent cloning of PCR products (LIC-PCR), 1990, Nucleic Acids Research 18 (20): 6069-6074. doi: 10.1093/nar/18.20.6069</li> | ||
</ol> | </ol> | ||
</div> | </div> | ||
Latest revision as of 00:29, 27 October 2012
The GoldenBricks: A fast cloning technique
for the PartsRegistry

Introduction
Context
Cloning is probably the most tedious and less rewarding task when engineering living entities through synthetic biology. With the development of the restriction enzymes and efficient DNA ligases, assembling small and large DNA pieces has become a common practice in biology, but as everyone is using their own sets of enzyme and assembly method, making large DNA constructs is still a challenge today. The first indisputable advance in the domain came with the invention of the first standardized DNA cloning method: the BioBricks [1]. The standard BioBrick format (BBF RFC 10) make possible to clone together all kind of genetic parts (called "bricks") using only four different enzymes: EcoRI, XbaI, SpeI and PstI. But the real power of BioBrick standardization lays in the possibility to build database made of these DNA pieces that anyone can be assembled through a standard method. The PartsRegistry today contains almost over 3,000 basic characterized biological parts [2] and is a sources of inspiration for thousands of biologists around the world.
If biobricks have opened new perspectives for engineering organisms and have proven their efficiency over a decade, their use is limited by the fact that the fragments can only be assembled one by one. On the top of this, the process is very difficult to automatize, because of the number of DNA purification step required. IGEMers today spend most of their time assembling genetic parts together, leaving little time for characterizing them and testing their systems.
Several cloning techniques capable of assembling multiple fragments at the time have been invented since the rise of the Biobrick format. The most popular one is probably the one known as the Gibson method [3] that can be used to assemble up to four fragments at the time on a regular basis. The Golden Gate technique [4] (>20 fragments at the time) and its new evolution, the MoClo [5] (47 fragments in two times reported) are leading the way of another kind of cloning technique based on type II restriction enzymes. More efficient than the Gibson cloning, the Golden Gate also have the huge avantage that it can assemble more parts at the time, no matter their size. Moreover, Golden Gate do not imply any DNA modifications, which noticeably reduce the probability of having a mutation at the ligation scar.
If these new techniques dramatically speed-up the assembly of DNA pieces, to our knowledge, no reported work has been carried on standardizing these methodologies. So far, biologists using Golden Gate engineer new overhangs and create a new library each time they are making a different construct.
From Golden Gate and BioBricks to GoldenBricks
The BioBricks are a collection of parts that can be assembled one by one in a generic way. Golden Gate is a technique that enable the assembly of several tens of parts simultaneously. Taking inpiration from both technique, the 2012 Evry iGEM Team invented and developped a new methodology called GoldenBricks, merging the power of the two techniques while keeping the compatibility toward the old RFC 10 BioBrick format and preserving the possiblilty to make standard GoldenGate assembly simultaneously with GoldenBrick assembly, using the same vectors.
GoldenBricks technique is a one-shot cassette construction using DNA parts coming either from a plasmid distribution or from PCR product, engaging 5, 7 and possibly more different parts. Moreover, GoldenBricks works for both eukaryotes and procaryotes DNA construction with the very same protocol. If a non classical assembly is required (as for testing the strength of a terminator) a new "split construction" method based on standard plasmids make possible to assemble parts in a non classical way.
Perspectives
Such technique offers great perspectives for the future of iGEM and of the PartsRegistry in general. First, it would make possible the fast and simple assembly of complete cassettes using less expensive equipment, less toxic chemical and less sequencing runs than the BioBrick assembly. Second, it would make fast and efficient cloning accessible for both researchers as well as for less experimented users, such as high school iGEMers or biohackers thanks to the simplicity of its protocol. Third, the creation of large expression cassettes using this method is cheaper and faster than synthesizing the construct at the present day, which would guaranty the interest for a DNA database compared to de novo synthesis for the years to come. And last, this method is a lot more easy to automatize unlike the RFC 10 biobrick format.
Theory
Requirements for the development of the new standard
In the continuation of what we wrote previously and in order to stay in the progression of the methodology we have tried in this work to stay as close as possible to the biobrick format, to keep GoldenBrick parts compatible with the RFC 10 standard assembly, while opening new perspectives to increase the speed and the easiness of cloning. These constraints imposed several requirements for this new standard.
First, the new assembly method should:
- Keep the compatibility with the current BBF RFC 10 standard
- Be compatible with a database approach
- Minimize the number of different prefixes and suffixes
- Use the standard registry plasmids and negative cloning control
To improve the cloning speed, we would also like to:
- Make one step Golden Gate cassette cloning possible
- Allow to check the construct with a single enzyme digestion
- Improve the compatibility with Gibson by removing the NotI site
- Provide a solution for non standard assemblies
As we will demonstrate, the GoldenBrick format we have established meet all these requirements. To understand on what basis we have created it, we are now going to explain in this section the principles that led the design of this format before we analyze the sequences of the GoldenBrick prefixes and suffixes and finally discuss what can be done with this technique.
A brief introduction on type II restriction enzymes
Golden Gate cloning, (also known as type II restriction enzyme cloning) rely on a specific familly of enzyme (known as the type IIS enzymes) very different from the ones we are used to. The characteristic of type IIS enzymes from the cloning point of view is that these enzymes cut the DNA outside their recognition locus, no matter the sequence of the place where they cut. A very small subtype of them can be used for cloning purpose, since they generate overhangs on one side of the recognition site only. The ones used for Golden Gate and MoClo cloning are BsaI and BbsI.
The fact that these enzymes cut outside its recognition site gives them two significant advantages:
- You can generate as many different overhang you want with a single enzyme within the same tube
- You can engineer your fragment so that it cannot be cutted again after it has ligated.
Thanks to this last point, it is possible to cut and ligate the DNA in the mean time within the same tube (BsaI works in the T4 ligase buffer), and do not require any DNA purification. For this reason, the ligation is achieved with a better yield and with less mutation than with traditional type II enzymes cloning methods.
Since the small BsaI fragment cut can re-ligate with the mother strand, the assembly is a lot more efficient if the tube is alternatively placed at 37°C, when the BsaI cuts and then placed at 16°C for a ligation time, using a thermocycler. This technique drive the equilibrium towards the product formation.
Analysis of a classical synthetic biology construction
A traditional construct in synthetic biology is made of the repetition of the following elements:
At the moment, GoldenBrick only allows the assembly of a single cassette at the time. In order to create the full system, the assembly of the different cassettes together have to be conducted afterwards with the biobrick assembly, Gibson assembly - that is very efficient to assemble long fragments - or ligation independant cloning [6]. Focusing closer on a single cassette, we can notice that we have four different kinds of junctions:
As we can notice on this picture, there is four different kinds of junctions in an expression cassette. In Golden Gate, the junctions are DNA overhangs that can be freely engineered thanks to the capacity of Type IIS enzymes to cut outside their recognition site, no matter what sequence is there. Therefore, we should engineer four different overhangs. The first overhang is dedicated to the ligation of the plasmid suffix with the promoter prefix (OH1). Similarly, the 5th one is dedicated to ligate the terminator suffix with the backbone prefix (OH5). In a bacterial device, the scar between the RBS and the gene (OH3) has to be precisely controlled because the distance between the two elements is critical for correct protein expression. This scar will remain the same than in the biobrick assembly, but it will be generated by a type IIS enzyme as for the other overhangs. Finally, the overhang that link the RBS prefix to the promoter suffix (OH2) and the protein suffix to the terminator prefix (OH4) have to be compatible to make the repetition of several {RBS-Protein} units in a single cassette (OH2=OH4) possible. We will see later how we can achieve control of this repetition.
To conclude, we have five different kinds of parts, which implies to create five different prefixes and five different suffixes with four different overhangs, which will help to assemble the five different DNA parts in the correct order.
In addition, we would like to generate a scar that can be digested by another enzyme, in order to control whether all the elements we intended to insert are present in the cassette or not, before sequencing it. As we will see later this element is critical for the selection of the clone that have inserted all the genes we want and avoid sequencing incorrect constructions. In order to generate such a scar, the overhangs 1, 2, 4 and 5 has been engineered in the middle of a BbsI site. When two pieces ligates, they recreate a BbsI restriction site, and the construct can be checked with a simple BbsI digestion.
One last condition to fulfill to get a RFC 10 compatible brick after GoldenBrick cloning is to leave no illegal restriction site inside the brick after the assembly of the cassette. However, we should keep the as similar as possible the BioBrick extensions around each elements leaving the four usual enzyme restriction sites, so that the brick can be still assembled using RFC 10 BioBrick method. The only drawback is that if several GoldenBrick parts are assembled with RFC 10 standard, they cannot be assembled with other GoldenBrick parts afterwards, because the BioBrick assembly will leave illegal BsaI site inside the casette.
The proposed new set of Golden Bricks extension
Having all these design principles in mind and meeting the requirements we have proposed, we designed the new GoldenBrick prefix and suffix with the following sequences:

This design fulfills all the requirements enumerated before and ensure that the different elements will be assembled in the correct order. We also generated a scar that can be digested afterwards with the BbsI enzyme, so that the correct insertion of the different elements can be checked afterwards by doing a single digestion. This also enable the polymerization of the {RBS-CDS} units, as we will discuss in the next paragraph.
Control over the polymerization
The only difference with a standard GoldenBrick protocol is the capacity for the RBS-protein cassette to polymerize. This polymerization capacity makes the power of the technique because it enables to make polycistronic mRNAs no matter what gene is inserted. However, a control mechanism is required in order to create polycistrons or when we want a single gene to be inserted in the cassette.
In order to achieve such a control, several parameters in the protocol can be ajusted to move the equilibrium from monocistrons to polycistrons. The first and the most obvious one is the stochiometry of the different elements. If the promoters and terminators are in large excess compared to the RBSs and the gene, the balance will be in favour of monocistrons. On the contrary, if the amount or RBSs and coding sequence in dominant, the cassette will tend to contain two or more genes inserted inside.
On the other hand, the number of cycle and the ligation time will also influence the polymerization. Polycistrons are not likely to appear when the number of cycle is small and the ligation time short, but they will become more and more important as we increase these parameters.
Most of the work carried on by our team is now focused on optimizing the protocol. We will release soon a set of standard protocols for mono and polycistrons optimized to reduce the heterogeneity of the polymerization length.
Assembly procedure
Assembling parts using the GoldenBrick method is a lot easier than for BioBricks and also five to ten times faster. We are now going to explain the procedure from the theoretical point of view. For the complete protocol, you should refer to the material and methods section.
Because it requires a very small amount of DNA, the plasmid source can be taken directely from a DNA distribution suck as the PartsRegistry distribution, without requiring the need of a DNA preparation before. Once the plasmids are resuspended, the experimentalist mix them together in a mix containing the BsaI enzyme and T4 ligase and put them in the thermocycler for 5 hours. At the end, the product can be directly transformed and plated. A first round of screening on the good polymerization length can be made using colony PCR with the standard VF2 and VR primers. If required, a second step of screening can be made after miniprepping using BbsI. Given the very small rate of mutation, only two clones with the good digestion or colony PCR signature can be sent for sequencing; they will contain the correct construct with a very high probability.
This technique however requires to screen an important number of clones because we don't have a total control over what is assembled inside the cassette, because of the freedom we left for the RBS-Protein entities to polymerize. We will show in the next paragraph how we can gain a relative control over it. To reduce the costs of screening, we encourage the use of colony PCR over mini-preping and digestion when possible.
Other advantages of GoldenBricks over any other standard cloning techniques
Non standard assembly and "split-construct" vectors
If you are willing to create a non standard assembly or to control very precisely RBS-protein pairing over a polycistrons, you might like to create the construct in two times and assemble them together in a second step. This is why we have created the "split-construct" vectors. The split-contruct vectors are similar to the goldenbrick vectors except that they contain the OH2-OH5 or the OH1-OH4 overhangs instead of the OH1-OH5 overhangs as the GoldenBrick plasmid.
These vectors enable the assembly of shorter casette containing only a promoter, and a serie of RBS-protein for the "split-construct" 1 vector, or RBS-protein terminator for the "split-construct" 2 vector. This way, if a non standard biobrick assembly has to be carried out in between the two pieces or if a long polycistron has to be created, they can be assembled using the standard EcoRI, XbaI, SpeI, PstI cloning or Gibson assembly, but several cloning step will be saved by using GoldenBricks compared to the standard assembly of the full cassette.

GoldenBrick make standard part DNA shuffling possible
Aside from the dramatic shortening of the cloning time, the decrease of the costs and the easiness of GoldenBricks over Biobricks, one of the most noticeable improvement is undoubtedly the possibility to make DNA shuffling using a standard format. Let us take the example of a complex system such as a toggle-switch, in which you would like to optimize the expression of the protein repression of its two components to place the system in the working area of the phase domain.
Using the Biobrick format, you will probably have to make tens of different constructs with different RBS to find a condition in which the toggle-switch will work. Using GoldenBricks, you can make all of them in a single tube, in a single step. Moreover, you may eventually find a way to screen them directly after the cloning avoiding the need of purifying and sequence all the clones that couldn't work.

Many other use of DNA shuffling can be found in the literature especially for gene expression optimization and screening for working enzymes. We expect that DNA shuffling over standard parts is going to find many applications in the future of iGEM projects.
GoldenBrick is still compatible with Biobrick format
If the situation requires the part to be assembled with the standard RFC 10 format, the new GoldenBrick parts can be assembled the very same way than BioBricks. The only limitation is that BioBrick assembly of GoldenBrick parts leave some scars containing the BsaI restriction site inside, and the produced parts cannot be reassembled with other parts using Golden Gate. No simple way of fixing this issue has been found.
GoldenBricked plasmids are compatible with standard Golden Gate assembly
The GoldenBricked plasmids can be used to assemble contructions using classical Golden Gate. This option has been tested in our team and it works fine. See the result section for more results on the use of GoldenBrick vectors with the construction of the IaaH-Pep2A-IaaM fusion protein.
GoldenBrick also assemble PCR products and shorten the assembly time
Someone that want to create a construct not having the time or the need of cloning his gene into a vector first can still assemble them using GoldenBrick. Although this practice wouldn't be encouraged for standards parts, it can become really useful when trying libraries of different genes in order to find a working one. GoldenBricked parts in plasmid and PCR GoldenBricked parts can be mixed together and assembled the very same way. The user will just have to take care to submit his working parts afterwards. We recommend to amplify new parts flanked by its GoldenBricks prefix and suffix, mix them with the GoldenGate assembly mix in one hand, and digest them in EcoRI and PstI on the other hand and ligate them in pSB1C3 for submitting the GoldenBricked part to the registry.
Classical GoldenGate can be carried out in the mean time than GoldenBrick assembly to create fusion proteins within the construct
GoldenGate in general and GoldenBricks in particular are very efficient to assemble fusion proteins. This is because one can design every overhang it wants and therefore fuse protein domains without leaving any scar. New fusion proteins can be created in the meantime they are assembled in the cassette, using an aditional scar designed by the experimentalist. We also recommand to submit the new fusion protein part separately to contribute to the registry.
Finally, GoldenBricks works for eukaryotes chassis as well as with prokaryotes
As we have seen all along this article, this design of GoldenBricks have been designed for procaryotes, but it can work very well for eukaryotes. In eukaryotes, promoters can include or not a 5'UTR in their sequence to enhance the protein expression. In the case there is no 5'UTR, we can use the same prefix and suffix than the prokaryote promoters and include the 5'UTR with the prokaryotes prefix and suffix of a prokaryotic RBS.
In order to make polycistrons, we can use an Intermediate Ribosome Entry Site (IRES), cloned between an prokaryote RBS prefix and suffix.
Finally, the polyA and the polyA capping cassette with a polymerase terminator can be flanked by the prokaryotes terminators sites. In the case one don't want to include a polyA capping, he can use the "split construct" 1 or in the case there is not terminator, the one on the plasmid will take care of stopping the RNA polymerase extension.
Results
Construction of the library
In order to test the GoldenBrick technique efficiently, and for easiness of screening and statistics, we have developed a set of GoldenBricked parts (see Material & Methods) with fluorescent proteins markers. We have created and submitted the following parts:
- K812050: A GoldenBricked version of pSB1C3 with J04450 as negative cloning control (status=sent)
- K812051: A GoldenBricked version of pSB1K3 with J04450 as negative cloning control (status=constructed)
- K812053: A GoldenBricked version of the strong RBS B0034 (status=sent)
- K812054: A GoldenBricked version of the RFP E1010 (status=sent)
- K812055: A GoldenBricked version of the terminator B0015 (status=sent)
- K812056: A GoldenBricked version of the pLac R0010 promoter (status=sent)
- K812057: A GoldenBricked of an sfGFP protein (status=constructed)
- K812058: A GoldenBricked of medium strenght RBS J61107 (status=constructed)
- K812059: A GoldenBricked of week RBS J61117 (status=constructed)
All the parts written here have been successfully constructed and sequenced and the ones marked as "sent" has been sent to the PartsRegistry.
Creation of a simple cassette
We have started experimenting on the GoldenBrick assembly method. For the moment, the results are not optimal so we are not publishing them yet. Better results are comming soon. For the moment, we have about 1% of correct clones.
Working with regular GoldenGate using GoldenBricks vectors
In order to assemble the fusion proteins we need for our project, we carried out two assemblies of fusion proteins using Golden Gate in our Golden Gated vectors. This paragraph reports the construction of the IaaH-Pep2A-IaaM fusion protein in the GoldenBrick plasmid K812050 using standard Golden Gate protocol. This protocol hadn't been optimized at the moment. However, out of 16 cloned on which we did colony PCR, 10 of them had the good size. We sequenced 4 of them and obtained the sequence expected.
This experiment demonstrates the possibility of doing GoldenGate in the GoldenBricks plasmids K812050 (and similarly in K812051) in a very efficient way.
Material and methods
Construction of the library
The GoldenBricked parts were constructed amplifying the corresponding biobricks using primers flanked by the appropriate GoldenBricks extensions, using Phusion (Thermo Scientific) or Q5 (NEB) DNA polymerase in a 30 cycle reaction. The PCR products were PCR purified and then digested using FastDigest® EcoRI and PstI (Thermo Scientific). The standard plasmid pSB1C3 (or pSB1K3) were digested similarely. Both vectors and inserts were PCR purified and then ligated using T4 DNA ligase (Thermo Scientific) before being transformed with home made competent cells (chemical or electroporation). Four white clones were inoculated and sequenced.
The either red or non red colonies were minipreped and then digested with BsaI for screening. All the clones that had the correct digestion pattern were sequenced and gave the correct sequence.
The RBSs were synthetized in the form of two matching oligos. A mixture of 10 µM of the two oligo was incubated at 98°C for 2 min and then cooled down to 4°C with a ramp of -1°C/s. They were then PCR purified, digested and PCR purified again before beeing ligated in pSB1C3.
Assembly protocol
The protocol given here hasn't been optimized yet. We are only starting the development of this new technology. It is given here as a matter of indication. Some optimized protocol will be available soon.
About 75 ng of each plasmid (more accurate ratios will be optimized) were mixed together with 2.5 units of BsaI enzyme (NEB) and 15 units of T4 DNA ligase (Thermo Scientific) in a total volume of 15 µL of standard T4 ligation buffer. The tubes were then placed in the thermocycler with the following steps:
- 37°C, 2 min (digestion)
- 16°C, 5 min (ligation)
- Goto 1, 50 times
- Option 1: if the construct does not contain any BsaI site: 50°C 5 min (reduce the background with a final digestion)
- Option 2: if the construct contains BsaI site: 16°C 2h (reconstitute the BsaI site)
- 80°C, 5 min (heat inactivation)
5 µL of the product was transformed with 50 µL of home made chemically competent cells and recovered for 1 hour. The cells were concentrated and then plated.
Perspectives for GoldenBricks and the PartsRegistry
When the complete protocols will be established, our team will submit a BBF RFC request for the registry. If the RFC get accepted and if the tenant of the PartsRegistry thinks this is a valuable format, we can think of migrating the registry to this new format. This can be done in two complementary ways: the first one is to ask every user to submit their new part in the new GoldeBrick format, and the second is to take advantage of the huge community working on the PartsRegistry and especially the iGEM competition. In order to migrate the already existing parts faster, we can add in the requirements for the gold medal to resubmit already exsiting parts in the new format. With an average of 200 teams in the years to come and asking each of them to resubmit two goldenbricked parts, we can cover the 2000 most useful parts within two or three years, because the teams will GoldenBrick the parts they are interested in for their assembly, so they will probably always re-submit more than two.
We believe that implementing such a method in the partsregistry will speed up the assembly process and give a new shine to this database that severely suffers from the gene synthesis and PCR based methods competition. This will leave more time for researchers and iGEMers for characterizing the parts, and the partregistry will also benefits from an increase of parts characterization, which is very beneficial to the community. Overhall, the idea of standardized parts that tends to loose its strength with the time will gain again in interest in the long run.
Conclusion
This year our team created a new cloning format for the PartsRegistry that dramatically speeds up the cloning time and opens new perspective. This includes simple protein fusion methods and DNA shuffling that were not possible using the current BioBrick format. At the moment, we are working very hard to demonstrate the possibilities of the technique and come up with efficient and reliable protocols. If the process proves its interest, we will submit an RFC to the PartsRegistry and propose the GoldenBrick progressively substitute to the current BioBrick format, which will renew the interest for people on standardized biological parts and standardized assembly over gene synthesis.
References:
- Knight TF, Idempotent Vector Design for Standard Assembly of BioBricks. 2003, Tech rep, MIT Synthetic Biology Working Group Technical Reports.
- Source: http://partsregistry.org/cgi/partsdb/Statistics.cgi
- Gibson D, One-step enzymatic assembly of DNA molecules up to several hundred kilobases in size. 2009, Nature Publishing Group: doi:10.1038/nprot.2009.77
- Engler C, Gruetzner R, Kandzia R, Marillonnet S, Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. 2009, PLoS ONE 4(5): e5553. doi:10.1371/journal.pone.0005553
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S, A Modular Cloning System for Standardized Assembly of Multigene Constructs, 2011, PLoS ONE 6(2): e16765. doi:10.1371/journal.pone.0016765
- Charalampos A & J. de Joung P, Ligation-independent cloning of PCR products (LIC-PCR), 1990, Nucleic Acids Research 18 (20): 6069-6074. doi: 10.1093/nar/18.20.6069
 "
"