Team:Wellesley HCI/Notebook/NicoleNotebook
From 2012.igem.org
| (14 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
/*actual content styles*/ | /*actual content styles*/ | ||
| - | body {width: | + | body {width: 900px; margin:auto;} |
#bu-wellesley_wiki_content {height:auto; line-height:100%;} | #bu-wellesley_wiki_content {height:auto; line-height:100%;} | ||
| Line 82: | Line 82: | ||
<link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM2012/css/Team.css"> | <link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM2012/css/Team.css"> | ||
| + | <link href='http://fonts.googleapis.com/css?family=Source+Sans+Pro:400,600' rel='stylesheet' type='text/css'> | ||
<style type="text/css">@import "http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/videobox.css";</style> | <style type="text/css">@import "http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/videobox.css";</style> | ||
| Line 89: | Line 90: | ||
<div id="bu-wellesley_wiki_content"> | <div id="bu-wellesley_wiki_content"> | ||
| - | + | <a href="https://2012.igem.org/Team:Wellesley_HCI"><img src="http://cs.wellesley.edu/~hcilab/iGEM2012/images/titleimage.png" width="300px" style="display:block; float:left;"></a> | |
| + | <!--Start NavBar--> | ||
<ul id="nav"> | <ul id="nav"> | ||
| - | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Team">Team</a></li> | + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Team">Team</a> |
| - | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Project_Overview">Project</a></li> | + | <ul> |
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Team">Team Members</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Notebook">Notebook</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Acknowledgement">Acknowledgement</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Tips_Tricks">Tips & Tricks</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Social">Fun</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Project_Overview">Project</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Project_Overview">Project Overview</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/SynBio_Search">SynBio Search</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/MoClo_Planner">MoClo Planner</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/SynFlo">SynFlo</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Downloads_Tutorials">Downloads & Tutorials</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| - | <li><a href=" | + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Human_Practices">Human Practices</a> |
<ul> | <ul> | ||
| - | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Methodology">Methodology</a></li> | + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Human_Practices">User Research</a></li> |
| + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Methodology">Methodology</a></li> | ||
<li><a href="https://2012.igem.org/Team:Wellesley_HCI/Safety">Safety</a></li> | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Safety">Safety</a></li> | ||
| - | + | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Outreach">Outreach</a></li> | |
| - | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Outreach">Outreach</a></li> | + | |
</ul> | </ul> | ||
</li> | </li> | ||
<li><a href="https://2012.igem.org/Team:Wellesley_HCI/Gold">Medal Fulfillment</a></li> | <li><a href="https://2012.igem.org/Team:Wellesley_HCI/Gold">Medal Fulfillment</a></li> | ||
| - | + | ||
| - | + | </ul> | |
| - | + | <!--End NavBar--> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<!--notebook goes here--> | <!--notebook goes here--> | ||
| Line 189: | Line 203: | ||
== Day 10: June 11, 2012 == | == Day 10: June 11, 2012 == | ||
| + | |||
| + | [[File:Brainstorm_activity.png|thumb|alt=One of our brainstorming boards|Brainstorming 1/3: eLab Notebook]] | ||
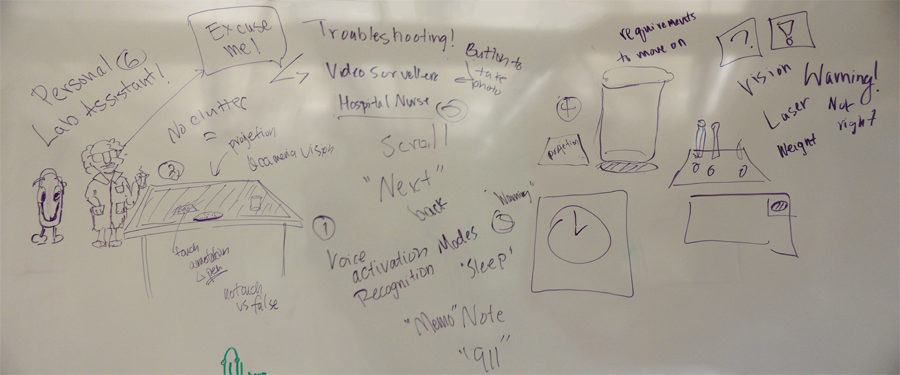
| + | [[File:Brainstorm_assistant.png|thumb|alt=One of our brainstorming boards|Brainstorming 2/3: Lab Assistant]] | ||
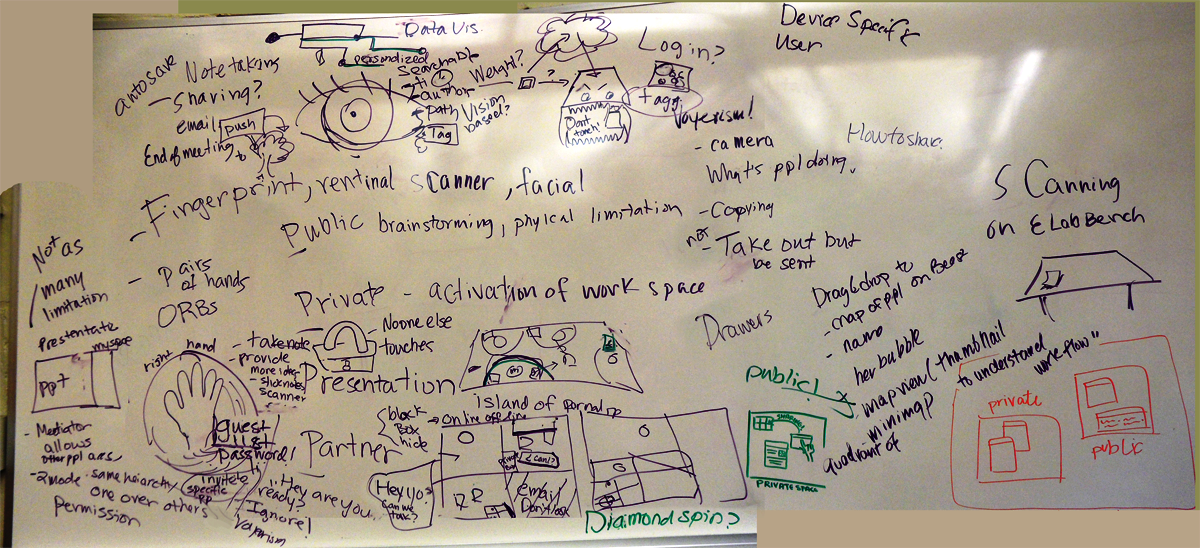
| + | [[File:Brainstorm_privatePublic_edit.png|thumb|alt=One of our brainstorming boards|Brainstorming 3/3: Private vs. Public space]] | ||
Today, each group presented their findings on the topics we had been assigned at the beginning of summer research. Between each presentation, we shared ideas that popped into our heads and devoted around 10 minutes to brainstorm how each of what had been researched could be integrated into our iGEM submission. | Today, each group presented their findings on the topics we had been assigned at the beginning of summer research. Between each presentation, we shared ideas that popped into our heads and devoted around 10 minutes to brainstorm how each of what had been researched could be integrated into our iGEM submission. | ||
| Line 201: | Line 219: | ||
== Day 12: June 13, 2012 - Day 13: June 14, 2012 == | == Day 12: June 13, 2012 - Day 13: June 14, 2012 == | ||
| + | [[File: IMG_20120613_144045.jpg|thumb|Veronica and I presenting our ideas about semantic search to the rest of the lab]] | ||
| - | + | Main brainstorming started today. In the morning, we set up a classroom such that each project had a different section. We stuck photos associated with our projects on the walls, and we wrote on post it notes and on the whiteboard of the ideas we currently had. When the BU team came over for the day, we went around to the different sections and had a huge group discussion for each project with the original subgroups leading the discussions. We talked about the possibilities of each project, any ideas pertaining to the project that popped out at other people, examples of similar projects done in the past, other things (e.g. platform applications) that can be incorporated into them (e.g. for the beast: DiamondSpin). We learned more about what the art project entailed and clarified the type of audience it would be aimed towards. Natalie Kuldell was able to stop by and gave us very useful input on what she thought of our current ideas for the eLab Notebook. Two things were made very clear: being able to search through the notebooks a scientist has accumulated throughout the years of their research would be a valuable tool; and, having the freedom to write whatever you want on the notebook is important, so we’ve started researching on styluses we could use. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | Main brainstorming started today. In the morning, we set up a classroom such that each project had a different section. We stuck photos associated with our projects on the walls, and we wrote on post it notes and on the whiteboard of the ideas we currently had. When the BU team came over for the day, we went around to the different sections and had a huge group discussion for each project with the original subgroups leading the discussions. We talked about the possibilities of each project, any ideas pertaining to the project that popped out at other people, | + | |
| - | + | ||
== Day 14: June 15, 2012 == | == Day 14: June 15, 2012 == | ||
| Line 214: | Line 228: | ||
== Day 15: June 18, 2012 == | == Day 15: June 18, 2012 == | ||
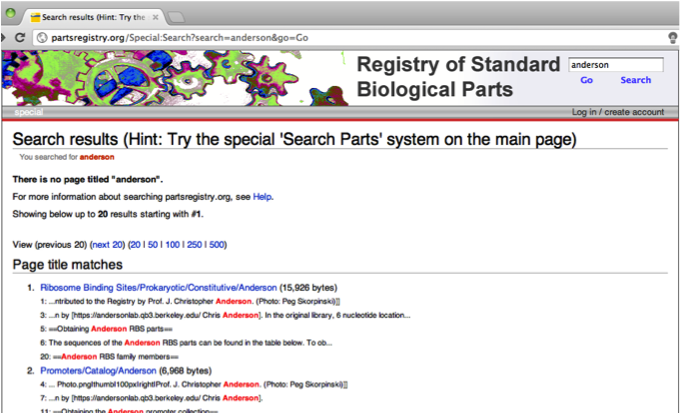
| + | [[File:Partsregistry-page.png|thumb|left|Sample search for partsregistry.org, which uses keyword search.]] | ||
Project status update. Currently finding it difficult to dump the database into XML and converting it into RDF because of the huge amount of data—might try to separate the site into smaller parts. While we are not very hopeful with our group’s part of the research because of the size of the database we’re trying to crawl through, we still haven’t exhausted all options and so are continuing with the research. Trying to look for anything similar to what we’re trying to do that had been done in the past, either by previous iGEM teams or not. Reading through the team abstracts from 2008, we weren’t able to find any team who had tried to convert the Parts Registry into RDF format or, in general, tried to create a search engine for it, semantic or otherwise. Veronica did however manage to find something called Knowledgebase of Standard Biological Parts, “a Semantic Web resource for synthetic biology” which managed to transform information from the Parts Registry to make it computable. This seems to be what we are essentially aiming to create, so investigating more on how the this resource was created will hopefully help us progress from our current rut. If we managed to create a structure for the Parts Registry, we would be able to access the information from individual teams’ notebooks. Trying to mine just part of the data (breaking it up into smaller collections), building own crawler, what else is necessary to customize our own engine. | Project status update. Currently finding it difficult to dump the database into XML and converting it into RDF because of the huge amount of data—might try to separate the site into smaller parts. While we are not very hopeful with our group’s part of the research because of the size of the database we’re trying to crawl through, we still haven’t exhausted all options and so are continuing with the research. Trying to look for anything similar to what we’re trying to do that had been done in the past, either by previous iGEM teams or not. Reading through the team abstracts from 2008, we weren’t able to find any team who had tried to convert the Parts Registry into RDF format or, in general, tried to create a search engine for it, semantic or otherwise. Veronica did however manage to find something called Knowledgebase of Standard Biological Parts, “a Semantic Web resource for synthetic biology” which managed to transform information from the Parts Registry to make it computable. This seems to be what we are essentially aiming to create, so investigating more on how the this resource was created will hopefully help us progress from our current rut. If we managed to create a structure for the Parts Registry, we would be able to access the information from individual teams’ notebooks. Trying to mine just part of the data (breaking it up into smaller collections), building own crawler, what else is necessary to customize our own engine. | ||
| Line 230: | Line 245: | ||
== Day 19: June 22, 2012 == | == Day 19: June 22, 2012 == | ||
| - | + | [[File:BIOFAB_Data_Access_Client-190520.png|thumb|BioFab's modular promoter library]] | |
Consuelo sent us another database from which to extract information to create a more complete parts datasheet: BioFab.com. BioFab’s projects are aimed to sharing information on the parts they themselves develop, but looking through how they organize this data gives us another idea of how our datasheets can look. Unfortunately, their database, C-dog is not searchable. They instead list out the and instead organize them by their name, construct, description, mean fluorescence per cell, and standard deviation. Choosing to organize them by name though is a bit frustrating. Each of their parts has the same prefix, “apFAB” followed by some number. Unfortunately, because they don’t precede these numbers with 0’s to ensure that 47 is not considered greater than 101 because the leading digit, 4, is greater than 1, then the order is not correct. However, I assume they provide information on standard deviation and mean fluorescence per cell because they are a major factor in helping biologists decide which part they need. We’ve started to finish up creating a more comprehensive data sheet using the information from the Parts Registry as listed by Linda in a document. However, because the pages are incomplete we are unable to extract some of the points. We still have to work on Web Crawlers for other databases, like BioFab and others, and customize the old Web Crawler for PubMed to extract information on parts instead of genes. | Consuelo sent us another database from which to extract information to create a more complete parts datasheet: BioFab.com. BioFab’s projects are aimed to sharing information on the parts they themselves develop, but looking through how they organize this data gives us another idea of how our datasheets can look. Unfortunately, their database, C-dog is not searchable. They instead list out the and instead organize them by their name, construct, description, mean fluorescence per cell, and standard deviation. Choosing to organize them by name though is a bit frustrating. Each of their parts has the same prefix, “apFAB” followed by some number. Unfortunately, because they don’t precede these numbers with 0’s to ensure that 47 is not considered greater than 101 because the leading digit, 4, is greater than 1, then the order is not correct. However, I assume they provide information on standard deviation and mean fluorescence per cell because they are a major factor in helping biologists decide which part they need. We’ve started to finish up creating a more comprehensive data sheet using the information from the Parts Registry as listed by Linda in a document. However, because the pages are incomplete we are unable to extract some of the points. We still have to work on Web Crawlers for other databases, like BioFab and others, and customize the old Web Crawler for PubMed to extract information on parts instead of genes. | ||
| Line 253: | Line 268: | ||
== Day 25: July 2, 2012 == | == Day 25: July 2, 2012 == | ||
| + | |||
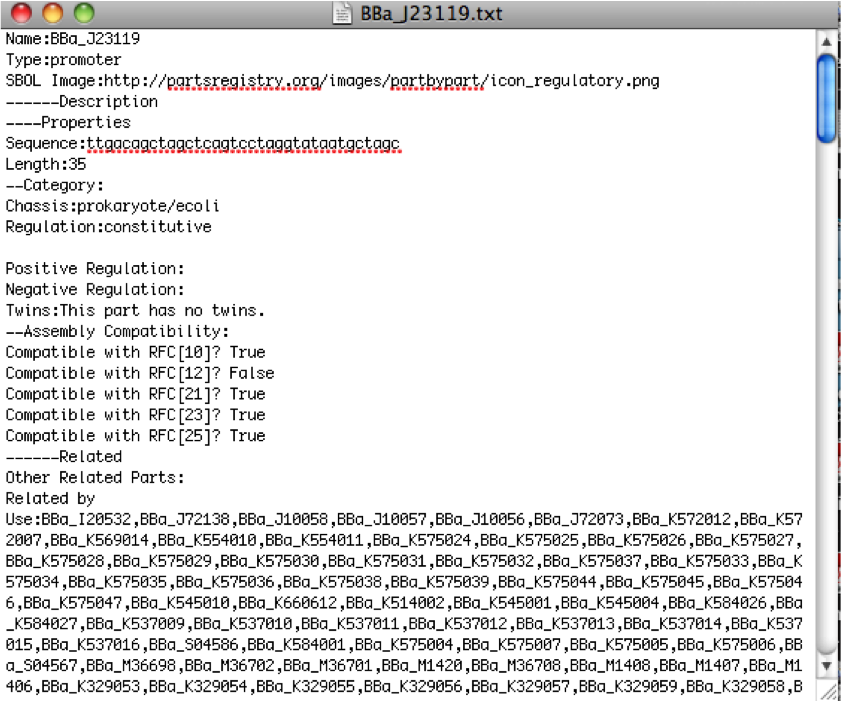
| + | [[File:Sample-Crawler-File.png|thumb|Sample .txt file produced by our RegList.cs and RegDataSheet.cs files]] | ||
Integrated PubMed Crawler -- instead of being able to find information on specific parts (because no results show up when one does that), just use a general text search and extract titles, authors, and abstracts. Worked on fixing the combination of the Results Page and Parts page parsers. | Integrated PubMed Crawler -- instead of being able to find information on specific parts (because no results show up when one does that), just use a general text search and extract titles, authors, and abstracts. Worked on fixing the combination of the Results Page and Parts page parsers. | ||
| Line 274: | Line 291: | ||
== Day 30: July 10, 2012 == | == Day 30: July 10, 2012 == | ||
| + | |||
| + | [[File:Blue-dripping-animation.jpg|thumb|left|The latest version of the dripping animation for one of the BioBrick options]] | ||
Kim and I were finally able to animate the finalized dripping images properly and the cubes appear with as they should when starting out. However, three main problems arose: one, when the E. Coli cube and the Colored cube are neighbors and the colored cube is tilted, the E. Coli cube runs the animation that should run on the Plasmid cube; two, after the dripping animation runs between the E. Coli and Plasmid cube, the Plasmid cube resets back to the original, blank image; and three, for some reason the order of the E. Coli images is wrong, so when the animation runs, the E. Coli looks like it's having a spasm attack. After speaking with Orit, I'm planning to start a major restructuring of the code then start working on the flipping functionality between the Plasmid and E. Coli cube. | Kim and I were finally able to animate the finalized dripping images properly and the cubes appear with as they should when starting out. However, three main problems arose: one, when the E. Coli cube and the Colored cube are neighbors and the colored cube is tilted, the E. Coli cube runs the animation that should run on the Plasmid cube; two, after the dripping animation runs between the E. Coli and Plasmid cube, the Plasmid cube resets back to the original, blank image; and three, for some reason the order of the E. Coli images is wrong, so when the animation runs, the E. Coli looks like it's having a spasm attack. After speaking with Orit, I'm planning to start a major restructuring of the code then start working on the flipping functionality between the Plasmid and E. Coli cube. | ||
| Line 286: | Line 305: | ||
== Day 34: July 16, 2012 == | == Day 34: July 16, 2012 == | ||
| + | [[File:IMG_20120717_112446.jpeg|thumb|left|SynFlo working with just three cubes]] | ||
Pilot test is on Thursday and presentation to the CS department is on Friday. Have to find a way to use more than just 3 cubes (for now, 5). To differentiate between each cube in the CubeSet so that code knows where to extract particular information (like color to use) I used instance variables to record the UniqueId of each cube (which is a permanent code of characters). I won't be able to use this method if the number of cubes in the CubeSet changes. If I try updating the UniqueId recorded depending on an interaction happening, then using multiple sets of cubes won't work. The main challenge is to ensure that the program works not when cubes are in sets of three, but any number above three. Still have to work on updating the UI of the surface application and have the textbox be updated with the color on the cubes -- am working on adapting the GnomeSurfer threading implementation. | Pilot test is on Thursday and presentation to the CS department is on Friday. Have to find a way to use more than just 3 cubes (for now, 5). To differentiate between each cube in the CubeSet so that code knows where to extract particular information (like color to use) I used instance variables to record the UniqueId of each cube (which is a permanent code of characters). I won't be able to use this method if the number of cubes in the CubeSet changes. If I try updating the UniqueId recorded depending on an interaction happening, then using multiple sets of cubes won't work. The main challenge is to ensure that the program works not when cubes are in sets of three, but any number above three. Still have to work on updating the UI of the surface application and have the textbox be updated with the color on the cubes -- am working on adapting the GnomeSurfer threading implementation. | ||
| Line 298: | Line 318: | ||
== Day 38: July 20, 2012 == | == Day 38: July 20, 2012 == | ||
| - | Today was presentation day. Worked on finishing up modifications to Parts Registry Web Crawler by having it pull more information on genes, specifically the type of gene as well as started work on optimizing search. When the program gathers the entire source code for the parts page, it is copied into each one of the big classes: BasicInfo, Description, Protocol, References, Promoter, RBS, Gene, and Terminator when only a fraction of the source code is needed to populate the instance variables in that code. Have to add more functionality to the cube, specifically the idea of contamination (where E. Coli can "contaminate" the colored and plasmid cubes) and, when neighboring another E. Coli, start an animation where each goes into the other's frame. Also have to work on lessening the memory space each animation takes up. | + | Today was presentation day to the rest of the Computer Science and Math Wellesley summer researchers. Worked on finishing up modifications to Parts Registry Web Crawler by having it pull more information on genes, specifically the type of gene as well as started work on optimizing search. When the program gathers the entire source code for the parts page, it is copied into each one of the big classes: BasicInfo, Description, Protocol, References, Promoter, RBS, Gene, and Terminator when only a fraction of the source code is needed to populate the instance variables in that code. Have to add more functionality to the cube, specifically the idea of contamination (where E. Coli can "contaminate" the colored and plasmid cubes) and, when neighboring another E. Coli, start an animation where each goes into the other's frame. Also have to work on lessening the memory space each animation takes up. |
== Day 39: July 23, 2012 == | == Day 39: July 23, 2012 == | ||
| + | [[File:2012-07-31_18.14.21.jpg|thumb|One of our first demo-able versions of SynFlo. Contamination of E. coli cubes and introduction of toxins have not been introduced yet]] | ||
| + | |||
Worked on optimizing search for MoClo Planner. While I was able to fix some of the bugs that lessened the amount of information extracted for particular parts, the search time for creating RegDataSheets for a list of seven parts actually increased from 33 seconds to 40 seconds. Did a task on the MoCloPlanner to help Kathy, Kara, and Nahum out with any glaring bugs that may have possible been in their program. Then worked on Sifteo for the rest of the day, mainly implementing the new animation where two E. Coli cubes, when they become neighbors, interact with each other. With the image set we previously had, the animation of an E. Coli looping around the screen was slow in the actual cubes compared to when I had tested it on the Computer Simulator ("Siftulator"). We had been afraid it was too quick, so Kim decided to make 5 copies for every one frame. Instead, it was too slow on the cubes because of its limited processing capabilities, so she just stuck with one copy of each frame, which kept the animation at the speed we wanted when we updated the cubes. Because we were able to free up the memory space, all the animation, including that of the surface, has become more fluid. I've installed the program on six cubes for the two demos happening tomorrow. | Worked on optimizing search for MoClo Planner. While I was able to fix some of the bugs that lessened the amount of information extracted for particular parts, the search time for creating RegDataSheets for a list of seven parts actually increased from 33 seconds to 40 seconds. Did a task on the MoCloPlanner to help Kathy, Kara, and Nahum out with any glaring bugs that may have possible been in their program. Then worked on Sifteo for the rest of the day, mainly implementing the new animation where two E. Coli cubes, when they become neighbors, interact with each other. With the image set we previously had, the animation of an E. Coli looping around the screen was slow in the actual cubes compared to when I had tested it on the Computer Simulator ("Siftulator"). We had been afraid it was too quick, so Kim decided to make 5 copies for every one frame. Instead, it was too slow on the cubes because of its limited processing capabilities, so she just stuck with one copy of each frame, which kept the animation at the speed we wanted when we updated the cubes. Because we were able to free up the memory space, all the animation, including that of the surface, has become more fluid. I've installed the program on six cubes for the two demos happening tomorrow. | ||
== Day 40: July 24, 2012 == | == Day 40: July 24, 2012 == | ||
| + | |||
| + | [[File:IMG_20120719_104236.jpeg|thumb|left|Kim presenting SynFlo to the first group.]] | ||
| + | [[File:IMG_20120719_112318.jpeg|thumb|I demonstrated the lab's Microsoft Surface to the same group.]] | ||
Two demos: first with eight kids and the second with four kids. I really liked Kim's presentation! The new way she presented and structured the information was definitely more engaging. The first half of the first group turned out well, mainly because everything ran smoothly and I was able to explain our project properly. The kids were also more enthusiastic and were very receptive towards the cubes, which they continuously played with throughout the demo. They especially liked introducing the E. Coli to the surface environment. During the second half however, both programs, especially the cubes' program, started going haywire. The animation would run but would keep blinking from Sifteo cube's default display to the dripping animation. Because the kids kept on running the animations without waiting for the others to end/near its end, the cube may have gotten overwhelmed. Overall however, when Kim asked each group leading questions in the end about what they learned about synthetic biology, they definitely absorbed more than the group from the pilot study. With the second demo, while the cubes were running smoothly, the kids were more mellow. They were still engaged with the demo but they were not as excited as the first half of the first group. Created separate text files for each type of part which contained a list of all the parts under that category including a keyword they can be used to filter through all the parts. Now, each type of part will contain all the parts in the text file as pre-loaded options. Instead of loading each part's information as soon as they appear as an option on a screen, the information is extracted from the registry only when the user clicks on the DataSheet option of that part, significantly lessening wait time. | Two demos: first with eight kids and the second with four kids. I really liked Kim's presentation! The new way she presented and structured the information was definitely more engaging. The first half of the first group turned out well, mainly because everything ran smoothly and I was able to explain our project properly. The kids were also more enthusiastic and were very receptive towards the cubes, which they continuously played with throughout the demo. They especially liked introducing the E. Coli to the surface environment. During the second half however, both programs, especially the cubes' program, started going haywire. The animation would run but would keep blinking from Sifteo cube's default display to the dripping animation. Because the kids kept on running the animations without waiting for the others to end/near its end, the cube may have gotten overwhelmed. Overall however, when Kim asked each group leading questions in the end about what they learned about synthetic biology, they definitely absorbed more than the group from the pilot study. With the second demo, while the cubes were running smoothly, the kids were more mellow. They were still engaged with the demo but they were not as excited as the first half of the first group. Created separate text files for each type of part which contained a list of all the parts under that category including a keyword they can be used to filter through all the parts. Now, each type of part will contain all the parts in the text file as pre-loaded options. Instead of loading each part's information as soon as they appear as an option on a screen, the information is extracted from the registry only when the user clicks on the DataSheet option of that part, significantly lessening wait time. | ||
| Line 320: | Line 345: | ||
== Day 46: August 1, 2012 == | == Day 46: August 1, 2012 == | ||
[[File:End-presentation.jpeg|thumb|alt=Preparing for the final presentation of the summer|Running my presentation by Orit]] | [[File:End-presentation.jpeg|thumb|alt=Preparing for the final presentation of the summer|Running my presentation by Orit]] | ||
| - | Tried having SynFlo work with Uppy, the Samsung SUR40 using Microsoft PixelSense. Fixed some bugs for reorientation of cubes, which is a lot more complicated than I had initially realized. Finished up the poster and started preparing for presentations. | + | Tried having SynFlo work with Uppy, the Samsung SUR40 using Microsoft PixelSense. Fixed some bugs for reorientation of cubes, which is a lot more complicated than I had initially realized. Finished up the poster and started preparing for presentations to the rest of the summer researchers, their parents, and professors. |
<!--notebook ends here--> | <!--notebook ends here--> | ||
Latest revision as of 18:58, 3 October 2012

Nicole's Notebook
Day 1: May 29, 2012
Research started today, so a majority of the day was dedicated to administrative details about the program. Otherwise, Veronica and I were assigned to research more about semantic search--what it was, its benefits, and whether or not it would be worthwhile to pursue this field in the context of synthetic biology. We will be presenting our findings near the beginning of June.
Day 2: May 30, 2012
For the majority of the day, I was learning how to code with C# on a Windows 8 Surface. It was easy to get lost on the more technical side and details of the coding, especially because the instructions seemed to assume you knew about the basic commands in C#. Other than that, it was easier to delve into the code without blindly copying down if it had been my first time. There were a couple of times where getting stuck and being unable to figure out the problem because of my lack of knowledge got frustrating, but because it was my first experience creating code to manipulate objects on a touch screen, I found the learning experience to be pretty amazing overall. Otherwise, I’ve been doing more reading on synthetic biology. It took me a while to read them since I hadn’t taken up Biology since the ninth grade, but I found the idea of engineering a new biological function interesting so I managed to complete most of the reading.
Day 3: May 31, 2012 - Day 4: June 1, 2012
I continued reading more about Semantic Search and its advantages compared to other types of searches as well as its disadvantages along with exploring different semantic search engines that are already available on the web, specifically for bioinformatic research (e.g. [http://bio2rdf.org], [http://distilbio.com], and [http://nlmplus.com]). A couple of papers also introduced me to the idea of incorporating other types of searches in Semantic Search. I learned a bit more about text mining, natural language processing (NLP), and ontology.
Day 5: June 4, 2012
Veronica and I continued reasearching on Semantic Search for the morning. The lab then headed over to Microsoft NERD for the second half of the day where we learned about their new beta project, a tablet with Microsoft 8. The interface is definitely engaging, what with their idea of using tiles and semantic zooming. I also like how transparent they’re trying to be with people who are trying to create applications for their story. Their idea of using a picture the owner can choose on their own as part of the password was really cool! I also like how they found a way to accommodate different finger sizes by enabling users to rescale the keyboard. All the different gestures to fully use the tablet however are not intuitive and takes some getting used to. It seemed much easier when Edwin was just demonstrating the product for us than when I actually tested out the tablet myself. Although, it is great how applications can now be made using HTML5 and JavaScript. I'm definitely going to start learning how to code in these languages before the summer is out.
Day 6: June 5, 2012
The lab met up in MIT and we spent the entire day learning more about synthetic biology. Professor Natalie Kuldell gave us a lecture in the morning and had us running experiments that afternoon to try applying what we've learned and get us exposed to the lab environment (which was particularly helpful for those of us who hadn't done Biology since 9th grade). Natalie was able to make the lecture engaging when talking about the basics of synthetic biology and its similarities with other engineering practices. Once she got more technical however it was more difficult for me to follow. As for the lab, first of all, I've definitely gained a newfound respect for people majoring in the biological sciences. Lab can be very tiring--and we were only in there for two hours when labs in Wellesley College courses usually take 4+ hours!
It was difficult to remember reasons behind you're doing the protocols you're doing especially if, like me, one is a beginner in the subject who's more immediately concerned with what the lab procedures mean by using a certain instrument or machine to complete a protocol. It was helpful that Natalie had led us into a discussions after the lab to reflect on what we just did and sort of touch upon the big picture, but I definitely felt like, by the end of the day, I learned more about lab processes rather than the material that motivated the experiments in the first place. Natalie highlighted how difficult it is to look for information when the researcher does not know what to look for--something that hopefully we'll be able to facilitate using the applications we'll be developing for iGEM and semantic search.
Day 7: June 6, 2012 - Day 9: June 8, 2012
Veronica, Casey, and I discussed our individual findings from the research we had done until this point and decided what we'd be presenting -- the basics of semantic search, ways we can implement semantic search in surface applications, and challenges of implementing semantic search. We also tried finding an already made API we could customize because creating our own semantic search engine is definitely not a feasible job for a few people in one summer.
We spoke with Consuelo and Orit about our current material for our presentation this coming Monday and found that we needed to research more on the feasibility of implementing a customized semantic search engine using databases like PubMed, iGEM, and the Parts Registry. We then finalized our presentation.
On Friday, we were able to speak with Professor Eni Mustafaraj. She went over a brief explanation of semantic search and how it is implemented. She gave us suggestions on ways we could have our own customized semantic search engine. Initially, we had thought of using the commercially-made engines that enabled users to modify it based on the databases they want the engine to use. Eni warned us however that we should search for semantic search engines that had been written about in a research/academic paper, and instead pointed us towards an open source project called Apache Jena. What we would have to do is somehow convert what we want into RDF format and from there figure out how we could implement the engine using Jena.
Day 10: June 11, 2012
Today, each group presented their findings on the topics we had been assigned at the beginning of summer research. Between each presentation, we shared ideas that popped into our heads and devoted around 10 minutes to brainstorm how each of what had been researched could be integrated into our iGEM submission.
I found the idea of the DiamondSpin (i.e. the Lazy Suzanne for platforms) really interesting. It has several functions but I found it to be most intuitive for the windows to shrink as they were dragged towards the center. Then, should a person at the other end of the Beast, for example, want to access it, they would just use the outer rim of the circle to rotate it such that the window’s orientation would be adjusted appropriately and it was closer to them. Then, they’d be able to drag it outside the circle and examine it. As for the idea of windows getting larger as they were dragged towards the center, while this may seem useful for presentations, considering that the circular form makes sense for a platform with people surrounding it like the Beast (change in orientations), then enlarging it in the middle to present it to people would keep it in one orientation. Consequently, the orientation may be readjusted enough times that it would become a distraction during the presentation.
After the presentations, we were split up into two different groups and each came up with five different big ideas for projects to pursue over the summer. I really liked the idea of having a graph, inspired by the idea of Semantic Search, that essentially tracked the person’s “trail of thought.” They begin with a main query that constitutes the middle bubble. Then, the succeeding nodes represent queries they made right after and/or related to the node it is attached to. If the person wanted to go back to change a query or start another trail, they’d be able to. There was also the idea of the personal lab helper (like the animated clip on Microsoft Word). The program would different between people by recognizing their irises or fingerprints to prevent confusion. The surface would be the lab table and through each protocol, if a certain tool was needed, the shape of the tool would appear on the surface and keep blinking until the person prepared the correct tool. A camera would be placed above the table and record while the researcher would be doing an experiment. There were also a lot of ideas for the eLab Notebook, like being able to zoom in to a protocol and annotate it but when you zoom out again, all that has changed is that the protocol is now either highlight or changed its color to red. The user would then know that they had annotations and there would be no clutter.
Day 11: June 12, 2012
The Wellesley Lab went over to Boston University. We were able to meet the two undergraduates who are part of the BU team. Traci presented more about Synthetic Biology and went into more details about parts names, what they did, and we learned more about what they are currently doing for the competition. Afterwards, three from the Wellesley team presented our research topics and the ideas that resulted from our brainstorming the day before. We then proceeded to brainstorm even more, gaining valuable insight from the BU team.
Day 12: June 13, 2012 - Day 13: June 14, 2012
Main brainstorming started today. In the morning, we set up a classroom such that each project had a different section. We stuck photos associated with our projects on the walls, and we wrote on post it notes and on the whiteboard of the ideas we currently had. When the BU team came over for the day, we went around to the different sections and had a huge group discussion for each project with the original subgroups leading the discussions. We talked about the possibilities of each project, any ideas pertaining to the project that popped out at other people, examples of similar projects done in the past, other things (e.g. platform applications) that can be incorporated into them (e.g. for the beast: DiamondSpin). We learned more about what the art project entailed and clarified the type of audience it would be aimed towards. Natalie Kuldell was able to stop by and gave us very useful input on what she thought of our current ideas for the eLab Notebook. Two things were made very clear: being able to search through the notebooks a scientist has accumulated throughout the years of their research would be a valuable tool; and, having the freedom to write whatever you want on the notebook is important, so we’ve started researching on styluses we could use.
Day 14: June 15, 2012
More research on pens, dump registry to XML and convert to RDF format for Apache Jena, transcribed audio files, terms and conditions of different databases like GoWeb, Google Scholar, etc.
Day 15: June 18, 2012
Project status update. Currently finding it difficult to dump the database into XML and converting it into RDF because of the huge amount of data—might try to separate the site into smaller parts. While we are not very hopeful with our group’s part of the research because of the size of the database we’re trying to crawl through, we still haven’t exhausted all options and so are continuing with the research. Trying to look for anything similar to what we’re trying to do that had been done in the past, either by previous iGEM teams or not. Reading through the team abstracts from 2008, we weren’t able to find any team who had tried to convert the Parts Registry into RDF format or, in general, tried to create a search engine for it, semantic or otherwise. Veronica did however manage to find something called Knowledgebase of Standard Biological Parts, “a Semantic Web resource for synthetic biology” which managed to transform information from the Parts Registry to make it computable. This seems to be what we are essentially aiming to create, so investigating more on how the this resource was created will hopefully help us progress from our current rut. If we managed to create a structure for the Parts Registry, we would be able to access the information from individual teams’ notebooks. Trying to mine just part of the data (breaking it up into smaller collections), building own crawler, what else is necessary to customize our own engine.
Day 16: June 19, 2012
Planned out the rest of the summer → starting to code/do technical feasibility checks. Initially, started research on web crawlers that pulled data from the Parts Registry website for the idea of having a semantic search engine for the Parts Registry among other things. However, because the process of trying to use Apache Jena is difficult, Conseulo suggested to use a Google Custom Search Engine through C# and have results pop up on a window. Managed to implement Google Custom Search Engine using C#, which is displayed in a DataGrid. Casey managed to code up a server which enables him to drag an image file from a tablet to a window and have it appear on the desktop computer. Linda managed to figure out a format for a data sheet for a part. Going to work on testing out how to search through PubMed and iGEM. Will check the quality of the custom search results compared to Parts Registry ‘Text Search’ results.
Day 17: June 20, 2012
Comparing Google Custom Search results between what shows up in the C# implementation and when testing out the custom search engine in the site, there is a significant difference. For one thing there are only 10 results that appear in the pop-up window of the C# code whereas there are 10 pages worth of results in the sample site. Moreover, the results do not show up in the same order, so now we are trying to figure out how to make the results that appear on the DataGrid window be more similar to how they should appear. To be able to search through PubMed, Consuelo provided us with the web crawler she coded for one of last summer’s projects. It definitely works with genes, so we’ll have to test out whether it works for phrases or other keywords. As for searching through the iGEM archive, we’ve added all previous iGEM sites to what the custom search engine will look through.
Unfortunately, we weren’t able to figure out how our problem with C# implement of Google custom search results in terms of order and amount, so we’ve decided to customize Conseulo’s previous web crawler for PubMed and modify it for the Parts Registry. So far, we’ve managed to find a URL scheme for text searches. We are learning the code behind her Web Crawler before we start editing it to better suit our needs. With the web crawler, we’ll also be handling the results that appear by going through the links and creating data sheets from the links to the different parts.
Day 18: June 21, 2012
Veronica and I managed to finish customizing part of Conseulo’s web crawler, so now we have a Web Crawler that goes through the search results of a query and extract the title of the pages, the links to said pages, and the number of results (which appears if the query is not too generic). Linda sent us a document listing the information she needs to be able to construct a data sheet, and we’ve come up with a structure for the classes needed to be able to extract the information in an organized manner and are currently working on the code.
Day 19: June 22, 2012
Consuelo sent us another database from which to extract information to create a more complete parts datasheet: BioFab.com. BioFab’s projects are aimed to sharing information on the parts they themselves develop, but looking through how they organize this data gives us another idea of how our datasheets can look. Unfortunately, their database, C-dog is not searchable. They instead list out the and instead organize them by their name, construct, description, mean fluorescence per cell, and standard deviation. Choosing to organize them by name though is a bit frustrating. Each of their parts has the same prefix, “apFAB” followed by some number. Unfortunately, because they don’t precede these numbers with 0’s to ensure that 47 is not considered greater than 101 because the leading digit, 4, is greater than 1, then the order is not correct. However, I assume they provide information on standard deviation and mean fluorescence per cell because they are a major factor in helping biologists decide which part they need. We’ve started to finish up creating a more comprehensive data sheet using the information from the Parts Registry as listed by Linda in a document. However, because the pages are incomplete we are unable to extract some of the points. We still have to work on Web Crawlers for other databases, like BioFab and others, and customize the old Web Crawler for PubMed to extract information on parts instead of genes.
Day 20: June 25, 2012
Until we get a response from our source about where to obtain more of the information we need to complete the data sheet, the code we have so far is essentially complete. Veronica and I managed to finish debugging and cleaning up our code (commenting, creating properties, etc.). This version of the crawler is only able to handle Parts pages. Because we were able to finish debugging before the end of the day, we try to use the HTML Agility Pack to try to parse through the other types of pages and try to extract all the content within the HTML and CSS style tags. Unfortunately, we ended up in a dead end because the resulting output was a mess due to the page’s formatting. Some pages just had headers and text, which was easy enough to interpret and if we had spent a bit more time we could have tried making the output relatively more readable for the reader. But, a lot of pages contained images, captions to these images, tables, and other representations of information, so the subsequent output was too much of a mess to read through. Instead, we’ll just have the Crawler process Parts pages, so next time we have to work on making sure that the rest of the code does just that (e.g. editing some classes to ensure that the lists containing the results to the search queries only contain links to Parts pages).
Day 21: June 26, 2012
Started to connect RegList.cs and RegDataSheet.cs via a method called search that takes a string (the query) as an argument. The first class parses through the search results page whereas the second crawls through the actual results pages. We need to connect it such that the Crawler invokes the RegDataSheet.cs for each link. A lot of troubleshooting needed to be done here, and difficulty was mostly caused because we hadn’t taken all cases into consideration. We’ve continued debugging by looking through the output of our classes and the actual information presented in the pages.
Day 22: June 27, 2012
Encountered problems with algorithm that parses through the search results page, so we spent the morning trying to solve it. Worked with Linda on connecting our back end of the MoClo Planner to the front end she is developing using HTML5 and JavaScript
Day 23: June 28, 2012
Reformatted the string returned by RegDataSheet in the order Linda wanted. Continue to find ways to extract other necessary information from the Parts Registry.
Day 24: June 29, 2012
Had a bit more difficulty with extracting the assembly compatibility but was able to complete the working code in the morning. Worked on making sure Parts Registry Crawler was commented and understandable. Started working on modifying Pub Med Crawler for our current purposes.
Day 25: July 2, 2012
Integrated PubMed Crawler -- instead of being able to find information on specific parts (because no results show up when one does that), just use a general text search and extract titles, authors, and abstracts. Worked on fixing the combination of the Results Page and Parts page parsers.
Day 26: July 3, 2012
Our summer interns from Framingham High School came in today! Two rising juniors, Maura and Madeleine. Linda and I started teaching them HTML and they started working on making their own website. In the morning, Casey and Consuelo held a tiny boot camp to make sure we were on the same page with coding and the Windows Application. Finished debugging crawlers and ensured that the output .txt file that the C# program creates is in the format Linda is able to work with. Created an executable file. Had to transfer it onto a new Console Application project so that Linda can access the code using a command line argument and through it populate her online data sheets. Have to read up on cgi-bins and how to create them for my code. Better commenting the code for Nahum and Kara to be able to use through getters.
Day 27: July 5, 2012
Was able to finish creating an executable/console application for the PartsRegBrowse code. Will continue work on coding up a CGI once Linda finishes the user interface for the front end. Debugged a bit more—modified for cases that had previously not been taken into account. Switched onto the Sifteo project so I talking with Kim about what she had currently coded up for the cubes and started learning about Sifteo API. We were able to make progress with a problem in the program. The colored cube can now drip into a neighboring plasmid cube! The next step is to be able to do that with whatever color the user decides on (i.e. red, yellow, or blue). Unfortunately, as we were trying to solve this next step to the problem and decided to restart from our last big progress (i.e. the dripping), we had found that that last update had not been saved onto the repository, so altering the code took longer than expected.
Day 28: July 6, 2012
Was able to get the animation to work with the other colors (blue and yellow). Met with Orit today to figure out the next step for Sifteo cubes so that we can have a demonstration by the end of the upcoming week. Have to include the E. Coli cube and code up how it appears alone and how it interacts with the others
Day 29: July 9, 2012
Started coding up the third (E. Coli) cube and trying to figure out how to continuously run the animation with the least amount, ideally without any, prompts. Currently having trouble with using the actual pictures for the dripping animation. There is more than twice the amount of frames for the finalized animation than there was for the animation created to test out the code. Not sure if it's because of the code (which should work regardless of the number of frames) or if it's because of the SiftDev/Siftulator App that images are not appearing on the simulated cubes. When Kim tested the finalized animation on the actual cubes, they worked with an error coming up (which we still have to figure out). Hopefully we'll be able to figure this out by tomorrow because it'd be great if I could concentrate on getting the E. Coli cube to work (including the main functionality behind it which is have it interact with the colored cube and, if said cube is flipped, color will transfer onto the E. Coli cube).
Really hoping to get all this done by Friday since Demo session with high school students is on Wednesday. Devoting Monday and Tuesday to debugging any more (hopefully minor) problems with the demo.
Day 30: July 10, 2012
Kim and I were finally able to animate the finalized dripping images properly and the cubes appear with as they should when starting out. However, three main problems arose: one, when the E. Coli cube and the Colored cube are neighbors and the colored cube is tilted, the E. Coli cube runs the animation that should run on the Plasmid cube; two, after the dripping animation runs between the E. Coli and Plasmid cube, the Plasmid cube resets back to the original, blank image; and three, for some reason the order of the E. Coli images is wrong, so when the animation runs, the E. Coli looks like it's having a spasm attack. After speaking with Orit, I'm planning to start a major restructuring of the code then start working on the flipping functionality between the Plasmid and E. Coli cube.
Day 31: July 11, 2012
I was able to recode one of the major classes and figured out how to solve the interaction problems I had found out about yesterday as well as the dripping animation issue and to have the Plasmid cube remain the same color even after the Plasmid and Colored cubes are no longer neighbors. Now, I'm working on the flip functionality between the Plasmid cube and the E. Coli cube but am currently running into problems.
Day 32: July 12, 2012
I spent the entire morning figuring out why the instance variables didn't seem to maintain the values that I had assigned them using other methods within the CubeWrapper class. It wasn't until I started using the StateMachine class to animate the Plasmid cubes that I figured out why the instance variables seemed to be resetting. Because I'm working with multiple cubes that interact with each other, assigning values for instance variables in one cube does not mean the same instance variables in all other CubeWrappers in the CubeSet will have the same value. Using the UniqueId property to differentiate between the Colored, Plasmid, and E. Coli cubes, I was able to code up the flipping functionality for all three colors. However, my code only works properly if there are three cubes that are part of the CubeSet--an assumption I'm going to have to fix since we'll definitely have at the very least 6 cubes connected.
Day 33: July 13, 2012
Consuelo and I started working on enabling communication between the Sifteo Cube program and a surface application she and Wendy had been working on. Because the surface application is on a continuous loop until it is closed, we are trying to thread it in the background.
Day 34: July 16, 2012
Pilot test is on Thursday and presentation to the CS department is on Friday. Have to find a way to use more than just 3 cubes (for now, 5). To differentiate between each cube in the CubeSet so that code knows where to extract particular information (like color to use) I used instance variables to record the UniqueId of each cube (which is a permanent code of characters). I won't be able to use this method if the number of cubes in the CubeSet changes. If I try updating the UniqueId recorded depending on an interaction happening, then using multiple sets of cubes won't work. The main challenge is to ensure that the program works not when cubes are in sets of three, but any number above three. Still have to work on updating the UI of the surface application and have the textbox be updated with the color on the cubes -- am working on adapting the GnomeSurfer threading implementation.
Day 35: July 17, 2012
Was able to adjust code so that more than just three cubes can be used (amount does not to be divisible by three either). Any colored cubes can introduce color into any Plasmid cube, which can introduce its color to any E. Coli cube. I'm not sure if it's only for the simulator, but each E. Coli cube cannot play animation (that's been started at different times) simultaneously. The previous E. Coli cube's animation has to finish before another E. Coli cube can run the animation. Updated Orit with current progress. We'll now be working on the "handshake" between the cubes and the surface, which should be finished by Thursday's pilot test with the high school students. Working on Friday's presentation (organizing the slides/making sure we know what to say).
Day 36: July 18, 2012
In the morning, high school counselors came to learn about our HCI lab as well as view the projects that we have been working on throughout the summer. For the rest of the day, I worked on refining the communication between the surface application (server) and the cubes (client). Goal: when an E. coli cube is placed onto the surface, in that same location the surface application creates the same colored E. coli which starts traveling around the screen. Each of the cubes have a byte tag on their bottom sides. The surface application detects these byte tags and establishes the connection, which the cube completes when it is flipped. This is to assign a byte tag to a specific cube, so only one cube at a time is able to be flipped. Ideally, once finished refining the code, the communication should be left open between the surface and the cubes and the surface has a local variable associated with the tag (i.e. a specific cube) that records its color. Because the connection remains open, the local variable is updated when the color of the cube changes. Moreover, when the surface application receives the color from the E. Coli cube, it sends back a confirmation string as well as a command to start the animation on the cube (of the E. coli leaving that cube).
Day 37: July 19, 2012
Consuelo and I were able to communicate what we wanted between cube and surface and so have the first working demo for Synfluo art project. Eight high schoolers came in for the demo today and I was happy with how cool they thought our installation was. They were very excited about playing with the Microsoft Surface. Kim started off with an introduction of synthetic biology then kids split into two groups, half of whom I showed the working demo to and the other with Kim for a discussion about risks involved. When I tried starting the demo however, although the Sifteo cubes worked, a problem arose when trying to communicate with the surface and it was because the internet connection went down. Meanwhile, Consuelo was able to distract them by showing them the Beast. It was great to see how enthusiastic some of the kids were in learning more about how the Beast worked, the back end of some of our programs, etc. The demo with the two groups of kids went smoothly after the initial incident. Had a meeting with Kathy and Kara about what information they need for the Registry Crawler. When Kara demo'd the MoCloPlanner for the video for tomorrow's presentation, I had just realized how slow it took the program to pop up results, so search optimization is something I'll also have to work on after extracting the information Kara and Kathy want.
Day 38: July 20, 2012
Today was presentation day to the rest of the Computer Science and Math Wellesley summer researchers. Worked on finishing up modifications to Parts Registry Web Crawler by having it pull more information on genes, specifically the type of gene as well as started work on optimizing search. When the program gathers the entire source code for the parts page, it is copied into each one of the big classes: BasicInfo, Description, Protocol, References, Promoter, RBS, Gene, and Terminator when only a fraction of the source code is needed to populate the instance variables in that code. Have to add more functionality to the cube, specifically the idea of contamination (where E. Coli can "contaminate" the colored and plasmid cubes) and, when neighboring another E. Coli, start an animation where each goes into the other's frame. Also have to work on lessening the memory space each animation takes up.
Day 39: July 23, 2012
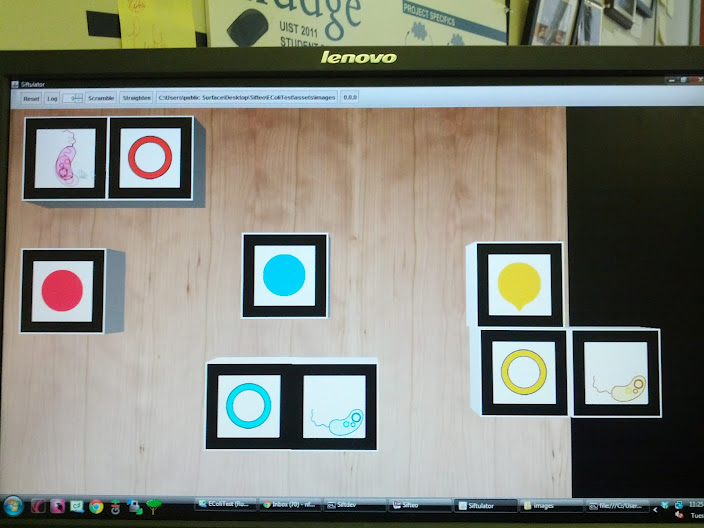
Worked on optimizing search for MoClo Planner. While I was able to fix some of the bugs that lessened the amount of information extracted for particular parts, the search time for creating RegDataSheets for a list of seven parts actually increased from 33 seconds to 40 seconds. Did a task on the MoCloPlanner to help Kathy, Kara, and Nahum out with any glaring bugs that may have possible been in their program. Then worked on Sifteo for the rest of the day, mainly implementing the new animation where two E. Coli cubes, when they become neighbors, interact with each other. With the image set we previously had, the animation of an E. Coli looping around the screen was slow in the actual cubes compared to when I had tested it on the Computer Simulator ("Siftulator"). We had been afraid it was too quick, so Kim decided to make 5 copies for every one frame. Instead, it was too slow on the cubes because of its limited processing capabilities, so she just stuck with one copy of each frame, which kept the animation at the speed we wanted when we updated the cubes. Because we were able to free up the memory space, all the animation, including that of the surface, has become more fluid. I've installed the program on six cubes for the two demos happening tomorrow.
Day 40: July 24, 2012
Two demos: first with eight kids and the second with four kids. I really liked Kim's presentation! The new way she presented and structured the information was definitely more engaging. The first half of the first group turned out well, mainly because everything ran smoothly and I was able to explain our project properly. The kids were also more enthusiastic and were very receptive towards the cubes, which they continuously played with throughout the demo. They especially liked introducing the E. Coli to the surface environment. During the second half however, both programs, especially the cubes' program, started going haywire. The animation would run but would keep blinking from Sifteo cube's default display to the dripping animation. Because the kids kept on running the animations without waiting for the others to end/near its end, the cube may have gotten overwhelmed. Overall however, when Kim asked each group leading questions in the end about what they learned about synthetic biology, they definitely absorbed more than the group from the pilot study. With the second demo, while the cubes were running smoothly, the kids were more mellow. They were still engaged with the demo but they were not as excited as the first half of the first group. Created separate text files for each type of part which contained a list of all the parts under that category including a keyword they can be used to filter through all the parts. Now, each type of part will contain all the parts in the text file as pre-loaded options. Instead of loading each part's information as soon as they appear as an option on a screen, the information is extracted from the registry only when the user clicks on the DataSheet option of that part, significantly lessening wait time.
Day 41: July 25, 2012
Planned abstract for ITS 2012 submission. Searched up how to upload Sifteo app onto the store. Finished grabbing lists of parts for Nahum to integrate for the new type of search/filter process for the MoClo Planner.
Day 42: July 26, 2012 - Day 43: July 27, 2012
Helped Kara and Veronica debug a new feature they implemented for primer designer of the MoCloPlanner. Wrote up abstract for submission of the SynFlo application to ITS 2012. Worked on the surface feature of SynFlo that simulates what occurs when E. coli is exposed to a contaminant. A new byte tag will be registered as the contaminant that causes the E. Coli encoded with the "red plasmid" to blur more than the other E. Coli. Ideally, when this application is complete, it is only when the E. Coli touches the byte tag that its state will change, but currently all red E. Coli change when the surface registers the tag. Finished implementations part of the abstract.
Day 44: July 30, 2012
Finished implementing the traveling E. Coli animation for all color combinations. Changed parser that constructed text files of most promoters/rbs/cds/terminators in registry that included parts' registry id and what type of part they were to include each part's common description name. Working on tag visualization of the surface application for SynFluo.
Day 45: July 31, 2012
Finished enabling reorientation for plasmid and color cube interaction and installed new program into cubes to test code out. Made it so that neighboring the Ecoli and Plasmid on any side of the other and flipping would run the animation and change the state of the cube. General preparations for poster session: Helped with some writing for the poster and taking pictures of the SynFluo demo.
Day 46: August 1, 2012
Tried having SynFlo work with Uppy, the Samsung SUR40 using Microsoft PixelSense. Fixed some bugs for reorientation of cubes, which is a lot more complicated than I had initially realized. Finished up the poster and started preparing for presentations to the rest of the summer researchers, their parents, and professors.
 "
"