Team:Kyoto/Notebook
From 2012.igem.org
(→March 6) |
(→Florigen) |
||
| Line 657: | Line 657: | ||
=Florigen= | =Florigen= | ||
| - | + | [[Team:Kyoto/Florigen/Note]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=Golden Gate Assembly= | =Golden Gate Assembly= | ||
Revision as of 05:18, 7 September 2012
Secretion
February 7
Preculture by_???
We started preculture at 12:10.
February 8
March 1
March 2
March 3
March 4
March 5
March 6
Restriction
| GFP | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 12 | 0.5 | 0.5 | 3 | 0.5 | 13.5 | 30 |
March 7
Electrophoresis1. 1kb ladder 2µL
2. pspA (PCR product)2.5µL + Loading Dye 0.5µL
3. GFP 30µL + Loading Dye 6µL
4. 1kb ladder 2µL
- The GFP was gel extracted on 3/6 and concentrated by Vacuum in 150µg/mL
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 3 | 3 | 3 | 9 |
| pspA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
at 4℃, for overnight
- torA→31.8ng/µL×3µL=95.4ng=0.529pmol
- pSB1C3→19.4ng/µL×3µL=58.2ng=0.042pmol
- pspA→49.3ng/µL×4µL=197.2ng=0.308pmol
- pSB1C3→19.4ng/µL×2µL=38.8ng=0.029pmol
March 8
Restriction
| pSB4K5 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 3 | 0.2 | 6.4 | 30 |
at 37℃ for 1 hour.
→Purification : 36.6ng/µL
Ligation
| Kil | pSB4K5 | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
at 4℃ for overnight
- Kil→37.7ng/µL×10µL=377ng=879fmol
- pSB4K5→36.6ng/µL×1µL=36.6ng=86fmol
Liquid culture
Lacp + pSB3C5 -1, 2
Transformation
| torA | pspA | competent cell | total |
|---|---|---|---|
| 1 | 0 | 10 | 11 |
| 0 | 1 | 10 | 11 |
We use commercially available competent cells in this time.
March 9
Restriction
| tatABCD | Xba1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
at 37℃ for 1hour
→purification : 75.0μg/mL (1.11 260/280 , 0.81 260/230)
Miniprep
lacP + pSB3C5 1 80.5μg/mL (1.78 260/280 , 2.00 260/230)
lacP + pSB3C5 2 107.2μg/mL (1.83 260/280 , 1.90 260/230)
Colony PCR
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 6sec
25cycles
Ligation
| tatABCD | constP J23107 | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
tatABCD : 227fmol
constP J23107 : 21fmol
Transformation
| pspA | torA | Kil | competent cell | |
|---|---|---|---|---|
| 1 | 1 | 0 | 0 | 10 |
| 2 | 0 | 1 | 0 | 10 |
| 3 | 0 | 0 | 1 | 10 |
Miniprep
4mL of plusgrow which had been cultured for overnight.
pSB4K5 : 80.5μg/mL
March 10
Screening PCR
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Then we did electrophoresis to confirm.
1.1kb ladder
2.kil (649bp)
3,4,5, pspA (969bp)
6,1kb ladder
1, 100bp ladder
2,3,4, torA signal
Restriction
| LacP-pSB3C5 | Spe1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 2 | 0.2 | 7.4 | 20 |
for 2.5 hours at 37℃
→purification 29.0ng/μL
| torA | Xba1 | Pst1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 2 | 0.2 | 7.4 | 20 |
for 2.5 hours at 37℃
→purification 91.8ng/μL
Ligation
| torA | Lacp-pSB3C5 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
torA : 929fmol
Lacp-pSB3C5 : 284fmol
for overnight at 4℃
March 11
Miniprep
We used 3μL of plus grow that we had cultured for overnight.
torA : 62.8ng/μL
Screening PCR
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
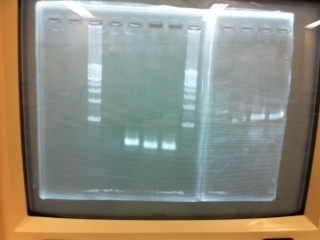
Electrophoresis
1. 1kb ladder
2〜11. pspA (969bp)
12. 1kb ladder
The results were shown as photograph in the right.
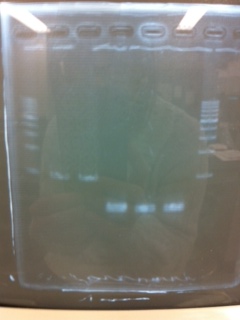
It seemed that there were shorter sample than expected sample, so we did electrophoresis with pspA which was product of PCR and pspA which had already cut with EcoR1 and Spe1.
1.1kb ladder
2. pspA (PCR product)
3, pspA (Eco, Spe)
4〜6, pspA (colony PCR)
7,1kb ladder
The results were shown as photograph in the right.
March 12
Transformation
| DT(1ng/μL) | DT(0.1ng/μL) | Kil | lacP-torA | MilliQ | competent cell | total |
|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 20 | 21 |
| 0 | 1 | 0 | 0 | 0 | 20 | 21 |
| 0 | 0 | 5 | 0 | 0 | 50 | 51 |
| 0 | 0 | 0 | 5 | 0 | 50 | 51 |
| 0 | 0 | 0 | 0 | 1 | 20 | 21 |
Restriction
| pspA | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.3 | 15.7 | 30 |
at 37℃ for 4 hours
→ We did purification and got 48.3ng/μL pspA.
Ligation
| pspA | pSB1C3 | MilliQ | Ligation High Ver.2 | total |
|---|---|---|---|---|
| 4 | 2 | 0 | 3 | 9 |
| 2 | 2 | 0 | 2 | 6 |
| 0 | 2 | 2 | 2 | 6 |
March 13
Miniprep pSB1C3 74.2µg/mL 1.65 (260/280) 1.31 (260/230)
March 14
Restriction
| pSB1C3 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 4 | 0.4 | 15.2 | 40 |
We did gel extraction and got 47.2ng/µL pSB1C3 but we did not cut out RFP.
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
| MilliQ | pSB1C3 | Ligation High Ver.2 | total |
| 4 | 2 | 3 | 9 |
at 16℃, for 1 hour
- torA : 0.707pmol
- pSB1C3 : 0.068pmol
Liquid culture
We cultured Lacp-pSB3C5 1,2,3,4,5 (CP tolerance)on culture with Amp.
→Only 4 which did not be cultured succeeded.
March 15
Liquid culture
We cultured Lacp-pSB3C5 6,7,8,9,10 on culture with ampicillin.
→6,8,9,10 were succeeded.
Restriction
| GFP | EcoR1 | Spe1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
| DT | EcoR1 | Xba1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
for 2 hours at 37℃.
Miniprep
pspA (pSB1C3) 40.5ng/µL
Ligation
| pspA | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 1.5 | 3 | 9.5 |
- pspA : 385fmol
- DT : 36fmol
Transformation
| J23107-tatABCD | DT (0.1ng/µL) | DT (0.01ng/µL) | pspA-DT | competent cells on 3/15 | total |
|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 20 | 22 |
| 0 | 2 | 0 | 0 | 20 | 22 |
| 0 | 0 | 2 | 0 | 20 | 22 |
| 0 | 0 | 0 | 2 | 20 | 22 |
Screening PCR
Kil, pspA and torA
| Quick Taq | Primer-r | Primer-f | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
March 16
Miniprep
torA (pSB1C3) 68.8ng/µL
Kil (pSB4K5) 92.7ng/µL
torA was red for some reason. We do not know why.
Colony PCR
| Quick Taq | Primer-r | Primer-f | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 6sec
→25cycles
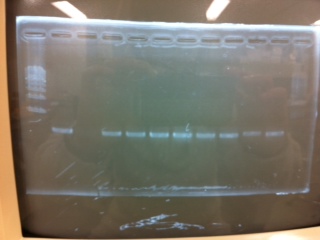
Electrophoresis
The results were shown as photograph in the right.
Checking Transformation Efficiency
competent cells that were made on March 15.
DNA : 0.02ng → 668 colonies Transformation Efficiency : 3.3×10^7
DNA : 0.2ng → 1739 colonies Transformation Efficiency : 8.7×10^6
Restriction
| pSB1C3 | EcoR1 | Spe1 | BSA | BufferM | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 0.3 | 3 | 0 | 6.3 | 30 |
| 5 | 0.2 | 0 | 0.2 | 0 | 2 | 12.6 | 20 |
at 37℃, for 2 hours.
We did gel extraction for product with EcoR1, Spe1. We got 46.1ng/µL pSB1C3.
| Kil(pSB4K5) | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
for overnight at 37℃.
We did this to confirm.
| pspA (pSB1C3) | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
for overnight at 37℃.
We did this to confirm.
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
| MilliQ | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
at 16℃, for overnight
- torA→767fmol
- pSB1C3→68fmol
March 17
Miniprep
J23107-tatABCD 72.7ng/µL
pspA-DT 50.5ng/µL
Checking the Insert
| J21037-tatABCD | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
Success.
| pspA-DT | EcoR1 | Pst1 | BSA | BufferH | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
Failed.
March 19
Restriction
| DT | EcoR1 | Xba1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did Gel extraction and got 17.2ng/µL of DT.
Ligation
| Kil | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 2 | 6 | 18 |
We did this for an hour at 16℃.
Restriction
| GFP | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 0.5 | 3 | 15.5 | 30 |
We did this for 4 hours at 37℃
Ligation
| pspA | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 5 | 5 | 5 | 15 |
| pspA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 4 | 1 | 3 | 8 |
We did these for an hour at 16℃.
- pspA (5µL)→377fmol
- DT→39fmol
- pspA (4µL)→339fmol
- pSB1C3→34fmol
Transformation
pspA-DT, pspA (pSB1C3), torA (pSB1C3) and GFP-DT
March 20
Screaning PCR
| Quick Taq | Primer-R | Primer-F | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
- pspA→○
- pspA-DT→○
- GFP-DT→○
- torA→×
- Kil-DT 6 of 8 sumples→○
| Quick Taq | VR | VF | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
- pspA→○
- pspA-DT→×
Restriction
| torA | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 0.3 | 3 | 16.1 | 30 |
| DT | EcoR1 | Xba1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did this for overnight at 37℃. And we did purification.
torA 34.2ng/µL
DT 28.0ng/µL
March 21
Miniprep
GFP-DT-1 : 77.7ng/µL
GFP-DT-2 : 67.6ng/µL
Restriction
| pSB1C3 | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
We did this for 3 hours at 37℃, and then we did gel extraction. We got 25.1ng/µL pSB1C3
| GFP-DT | EcoR1 | Pst1 | Xba1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0 | 0.2 | 2 | 12.6 | 20 |
| 10 | 0.2 | 0 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did this for 3 hours at 37℃, and then we did Purification. We got 30.7ng/µL GFP-DT.
Ligation
| torA | pSB1C3 | pspA | DT | GFP-DT | Ligation High Ver.2 | total | |
|---|---|---|---|---|---|---|---|
| 1 | 4 | 1 | 0 | 0 | 0 | 3 | 8 |
| 2 | 0 | 1 | 7 | 0 | 0 | 4 | 12 |
| 3 | 0 | 0 | 5 | 3 | 0 | 4 | 12 |
| 4 | 3 | 0 | 0 | 0 | 5 | 4 | 12 |
We did this for an hour at16℃.
March 22
PCR
We did PCR to amplify torA_signal that was product of PCR at March 5 with redesigned primers.
| Buffer | dNTPs | MgSO4 | Primer-F | Primer-R | Template | MilliQ | KOD plus neo | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 32.5 | 1 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 10sec
→30cycles
→Purification 110.7ng/µL
Restriction
| torA | EcoR1 | Spe1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
Ligation
| torA | pSSB1C3 | pspA | DT | GFP-DT | Ligation high | total | |
|---|---|---|---|---|---|---|---|
| 1 | 4 | 3 | 0 | 0 | 0 | 4 | 11 |
| 2 | 3 | 0 | 0 | 0 | 3 | 3 | 9 |
| 3 | 0 | 0 | 5 | 5 | 0 | 5 | 15 |
- torA (4µL)→512fmol
- pSB1C3→54fmol
- torA (3µL)→384fmol
- GFP-DT→36fmol
- pspA→377fmol
- DT→65fmol
March 23
Screening PCR
torA (pSB1C3), torA-GFP-DT and pspA-DT
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
E.coli in liquid culture that had pspA(pSB1C3) also expressed RFP.
Restriction
| pspA | EcoR1 | Spe1 | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.3 | 0.3 | 3 | 0.3 | 21.1 | 30 |
And then we did ethanol precipitation
Ethanol precipitation
pspA 11.5ng/µL.
Miniprep
Lacp+pSB3C5-8 77.9µg/mL
Lacp+pSB3C5-10 69.0µg/mL
Kil+DT-4 58.0µg/mL
Kil+DT-7 15.2µg/mL
Restriction
| Lacp+pSB3C5-8 | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
| Kil+DT-4 | Xba1 | Pst1 | Buffer2 | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1.5 hours
And then we did Gel extraction.
Gel Extraction
Lacp+pSB3C5-8 40.4µg/mL
Kil+DT-4 26.8µg/mL
March 26
Miniprep
torA(pSB1C3) 18.5[ng/µL]
torA-GFP-DT 20.0[ng/µL]
Restriction
| torA(pSB1C3) | EcoR1 | Pst1 | BufferH | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 2 | 0.2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 2 | 0.2 | 12.6 | 20 |
| torA-GFP-DT | EcoR1 | Xba1 | Pst1 | BufferH | BufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 0.2 | 0 | 0.2 | 2 | 0 | 0.2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0 | 2 | 0 | 0.2 | 12.6 | 20 |
| 20 | 0 | 0.2 | 0.2 | 0 | 3 | 0.3 | 6.3 | 30 |
We did Gel extraction and then got ??? 28.7[ng/µL]
Ligation
| Lacp (pSB3C5) | torA-GFP-DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
- Lacp : 22fmol
- torA-GFP-DT : 197fmol
| pspA | pSB1C3 | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
| pspA | DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
- pspA : 180fmol
- pSB1C3 : 18fmol
- DT : 16fmol
| LacP(pSB3C5) | Kil-DT | Ligation High Ver.2 | total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
for 2 hours at 16℃
Transformation
| Lacp-Kil-DT | competent cell | total |
|---|---|---|
| 1 | 10 | 11 |
March 27
Miniprep by
We retryed miniprep of torA(pSB1C3).
We got torA and its concentration was 39.3[ng/µL].
Transformation
| Name | Well | Sample | Competent Cells | Total | Plate | Colony |
|---|---|---|---|---|---|---|
| BBa_K117004 | 14J(2011 plate2) | 5 | 20 | ? | ? | ? |
We added 100[µL] of culture medium before we started culturing the E.coli.
Screening PCR
| Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Lacp-torA-GFP-DT 1, 3, 5, 6, 8, 9 was successful.
pspA-DT was failed.
E.coli that had pspA(pSB1C3) did not make any colony.
Lacp-Kil-DT 1,2,3,4,5,6,7,8 was failed.
Liquid culture
Lacp-torA-GFP-DT
Florigen
Golden Gate Assembly
August 10
Golden Gate Assembly method This method helps us to constract some genes quickly and we can design the order of constractions. We are trying to construct a gene cycle composed of 6 genes( pSB1A3, lacI, GFP, RFP, GFP, DT). After this experiment is confirmed to work, we will so some experiment to check how the numbers of promorters influences the strength of transcription.
 "
"