Team:Groningen
From 2012.igem.org
| Line 33: | Line 33: | ||
padding-right:10px; | padding-right:10px; | ||
padding-left:10px; | padding-left:10px; | ||
| - | margin: | + | margin: 0 auto; |
background-color: transparent; | background-color: transparent; | ||
background-color: rgba(0,0,0,0.3); | background-color: rgba(0,0,0,0.3); | ||
| Line 43: | Line 43: | ||
<body> | <body> | ||
| - | |||
<div class="cte"> | <div class="cte"> | ||
| + | <div class="ctd"> | ||
<l1 >Foodwarden</l1><br> | <l1 >Foodwarden</l1><br> | ||
</div> | </div> | ||
Revision as of 13:32, 22 August 2012








|
The Food Warden: it's rotten and you know it!
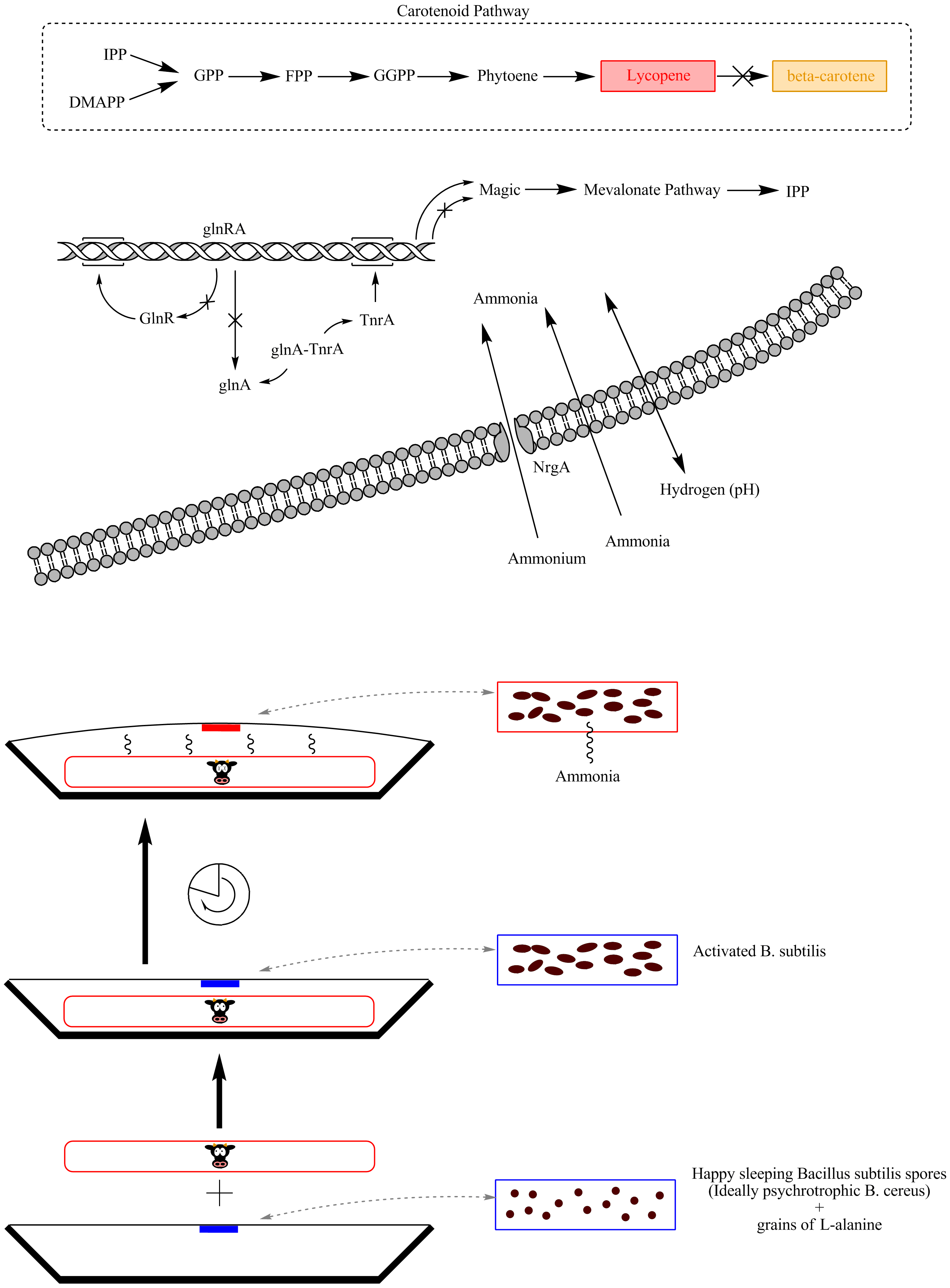
Our goal is to build a system to sense meat spoilage. When a package of meat is saved after opening, a closed sticker containing Bacillus subtilis spores is activated by breaking the middle compartment, similar to activating a glow-in-the-dark stick, whereby alanine and water are mixed with the spores to cause germination. When the germinated microbes come in contact with volatiles from rotting meat, a pathway is activated which results to the production of a pigment. When this happens, the consumer knows that he has to throw away the food. We use two strategies to make Bacillus subtilis react to rotten meat volatiles. The first involves TnrA, a transcription regulator involved in nitrogen metabolism. The picture below summerizes this goal pathway. The other strategy comprises the identification of new promoters reacting to rotten meat volatiles. In both cases, the promoter will be combined to a pigment production pathway. Besides that, iGEM Groningen 2012 aims to identify the volatiles that cause the promoter induction by using Gas Chromatography and Mass Spectrometry. A detailed description of our project plan can be found on the project page.
|
|
|
| Fig. h1. Overview of the functional reaction pathway for B. salus carnis. |
 "
"