Team:Bielefeld-Germany/Project/Appoach
From 2012.igem.org
KevinJarosch (Talk | contribs) (→Isolation and Generating of new BioBricks) |
KevinJarosch (Talk | contribs) (→Isolation and Generating of new BioBricks) |
||
| Line 35: | Line 35: | ||
For more information about the Organisms [[Team:Bielefeld-Germany/Project/Background/Organisms |click here]]. | For more information about the Organisms [[Team:Bielefeld-Germany/Project/Background/Organisms |click here]]. | ||
| - | |||
| - | + | These BioBricks are used to design new plasmids and vector-systems. The diffenten parts to realise a functional Plasmid btw. Shuttle-Vector-System are shwon in the following table: | |
| - | + | ||
| + | {| Parts of the ''E.coli'' Expressionvector and the ''P.Pastoris'' Shuttle-Vector!!|- |- | ||
| + | |T7 Promotorregion|| AOX Promotor|} | ||
| + | {|Plasmidcellspacing=0, Shuttle-Vector=0,} | ||
| + | !Plasmid Characteristic | ||
| + | !Shuttlevector Characteristics | ||
| - | The | + | |- |
| + | |rowspan="5"|Expression | ||
| + | |- | ||
| + | |T7 Promotorregion | ||
| + | |AOX-Promotor | ||
| + | |- | ||
| + | |His-Tag-Sequence | ||
| + | |alpha-factor | ||
| + | |- | ||
| + | |Chloramphenicol Resistant | ||
| + | |Zeocin Restitent | ||
| + | |||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | The Choice which expression system is used depends on the folding and glycosylation of the different laccases. | ||
| + | The genetically modified organisms are used to produce different laccases. Further down the line, these laccases are isolated and purified. | ||
| + | |||
| + | |||
| + | == Determining Activity and Potential of The Different Laccases == | ||
| + | |||
| + | The next step to generate a effeciantally filter system is | ||
Revision as of 15:53, 15 August 2012
Approach
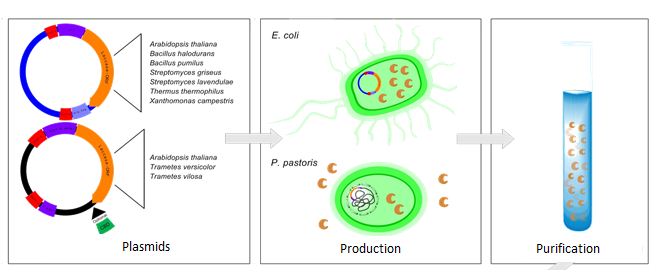
The conventional methods in sewage treatment plants are unable to treat waste water sufficiently because the most frequently used micro contaminants like estrogen, Bisphenol A, Dicolfenac etc. are very difficult to break down. The goal of Bielefeld’s iGEM team is to develop a biological filtersystem using immobilized laccases to purify municipal and industrial wastewater from synthetic estrogens and other aromatic compounds. Laccases are copper-containing oxidase enzymes found in many organisms, and one of their properties is the ability to break down a wide range of aromatic and phenolic compounds. For this purpose, genes of various bacterial and eukaryotic laccases should be isolated and expressed in Escherichia coli and Pichia pastoris. The choice of the expressionsystem depends on the glycosylation status of the enzyme.
Isolation and Generating of new BioBricks
The first step for Step of our project is to isolate the specific gene sequences and to generate new BioBricks for the iGEM competition. The Laccases of the following organisms are isolated:
bacterial laccases:- Escherichia Coli
- Baccilus Halodurans
- Baccilus Pumilus
- Streptomyces griseus
- Streptomyces lavendulae
- Thermus thermophilus
- Xanthomonas campestris
Eukaryota laccases:
- Arabidopsis thaliana
- Tramestes versicolor
- Trametes villosa
For more information about the Organisms click here.
These BioBricks are used to design new plasmids and vector-systems. The diffenten parts to realise a functional Plasmid btw. Shuttle-Vector-System are shwon in the following table:
| T7 Promotorregion | }
</center> The Choice which expression system is used depends on the folding and glycosylation of the different laccases. The genetically modified organisms are used to produce different laccases. Further down the line, these laccases are isolated and purified.
Determining Activity and Potential of The Different LaccasesThe next step to generate a effeciantally filter system is |
 "
"