1) ccm transformation
competent E coli were transformed with ccm plasmids
2)Citrobacter and E. coli growth on various sugars II
The reason there was growth on all plates previously was because the casamino acids that we added to our previous M9 plates acted as a carbon source. M9 plates were poured(omitting the casamino acids this time) without any carbon source. Small wells were cut in the middle of the plates and 150 μl of 20% sugar solutions (glucose, sucrose, lactose, glycerol) were added to each well. To some plates, powdered sucrose or glucose was added instead. The plates were then streaked with the same four strains as before ( E. coli, E. coli + sucrose hydrolase gene, Cf NCIMB and Cf SBS197) and incubated overnight at 37C.
Template:Team:Edinburgh/Notebook/Week05
From 2012.igem.org
(Creating a page for week 5) |
m |
||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:Edinburgh/header}} | ||
{{Team:Edinburgh/css/wiki-edit-structure.css}} | {{Team:Edinburgh/css/wiki-edit-structure.css}} | ||
| - | {{Team:Edinburgh/css/notebook | + | {{Team:Edinburgh/css/notebook-style.css}} |
| + | {{Team:Edinburgh/css/article.css}} | ||
| + | |||
| + | {{Team:Edinburgh/notebook-navigation}} | ||
<html> | <html> | ||
| - | <div id="week05"> | + | <style type="text/css"> |
| - | + | #week05 a, | |
| - | + | #week05 a:hover, | |
| - | + | #week05 a:visited, | |
| - | </div> | + | #week05 span{ |
| - | <div class=" | + | color: #7f7f7f; |
| - | <p>Posted on | + | font: bold 14px/18px tahoma, calibri, sans-serif; |
| - | </div> | + | } |
| - | <div class=" | + | #week05 .navigation-description{ |
| - | <p> | + | color: #ccc; |
| - | </p> | + | font: normal 12px/14px tahoma, calibri, sans-serif; |
| - | </div> | + | } |
| - | </div> | + | </style> |
| - | </div> | + | <div id="page-content"> |
| + | <div class="article" id="w05-23-07"> | ||
| + | <div class="article-title"> | ||
| + | <p><a name="week05-23-07">Week 5, 23.07.2012, Monday</a></p> | ||
| + | </div> | ||
| + | <div class="article-details"> | ||
| + | <p><Posted on 1/8/2012></p> | ||
| + | </div> | ||
| + | <div class="article-text"> | ||
| + | <p> | ||
| + | <b>1) ccm transformation</b><br/> | ||
| + | competent E coli were transformed with ccm plasmids<br/><br/> | ||
| + | <b>2)Citrobacter and E. coli growth on various sugars II</b><br/> | ||
| + | The reason there was growth on all plates previously was because the casamino acids that we added to our previous M9 plates acted as a carbon source. M9 plates were poured(omitting the casamino acids this time) without any carbon source. Small wells were cut in the middle of the plates and 150 μl of 20% sugar solutions (glucose, sucrose, lactose, glycerol) were added to each well. To some plates, powdered sucrose or glucose was added instead. The plates were then streaked with the same four strains as before ( E. coli, E. coli + sucrose hydrolase gene, Cf NCIMB and Cf SBS197) and incubated overnight at 37C.<br/><br/> | ||
| + | </p> | ||
| + | </div><!-- /article-text --> | ||
| + | </div><!-- /article --> | ||
| + | <div class="article" id="w05-24-07"> | ||
| + | <div class="article-title"> | ||
| + | <p><a name="week05-24-07">Week 5, 24.07.2012, Tuesday</a></p> | ||
| + | </div> | ||
| + | <div class="article-details"> | ||
| + | <p><Posted on 1/8/2012></p> | ||
| + | </div> | ||
| + | <div class="article-text"> | ||
| + | <p> | ||
| + | <b>1) ccm transformants subculture</b><br/> | ||
| + | ccm transformed cells that lost RFP were subcultured. <br/><br/> | ||
| + | <b>Citrobacter and E. coli growth on various sugars III</b><br/> | ||
| + | A final test involved adding the various sugars to the M9 medium in the bottle as opposed to either adding it to the plate before pouring the agar or adding them into the well on the plate. 5x100ml M9 medium bottles were prepared as before and autoclaved. The sugars (1ml) and thiamine hydrochloride (3.4 ml) were added to the bottles prior to the agar getting poured, with two bottles having no sugar added to them. After the agar had set, the plates were inoculated as before. One of the no sugar plates had solid citrate added to its middle, as before with the solid glucose and sucrose, to test the growth of Cf on this medium that gives it its name.<br/><br/> | ||
| + | </p> | ||
| + | </div><!-- /article-text --> | ||
| + | </div><!-- /article --> | ||
| + | <div class="article" id="w05-25-07"> | ||
| + | <div class="article-title"> | ||
| + | <p><a name="week05-25-07">Week 5, 25.07.2012, Wednesday</a></p> | ||
| + | </div> | ||
| + | <div class="article-details"> | ||
| + | <p><Posted on 1/8/2012></p> | ||
| + | </div> | ||
| + | <div class="article-text"> | ||
| + | <p> | ||
| + | <b>1) Liquid cultures preparation</b><br/> | ||
| + | subcultures of ccm transformed cells were used to inoculate 3ml LB; samples were left to grow overnight<br/><br/> | ||
| + | <b>2) Primers preparation </b><br/> | ||
| + | Stock solution (500 pmol/ul, solution in EB) and working solution (10 pmol/ul 1 in 50 silution of stock solution in water) of primers were prepared for nfsl, cymA, mtrA and mtrCAB gene primers. <br/><br/> | ||
| + | <b>3) Kod PCR of Shewanella </b><br/> | ||
| + | Kod Pcr was performed for potential Shewanella oneidensis strain using 3 sets of primers: cymA, mtrA and mtrCAB. Gel analysis indicated no results.<br/><br/> | ||
| + | <b>4)Newcastle!</b><br/> | ||
| + | Eugene, Oscar and Réka travelled long and far to Newcastle to have the nice people there sequence our two Cf strains with their new Ion torrent sequencing suite. We also got the grand tour of their Centre for Bacterial Cell Biology, located in the new Baddiley-Clark Building, with detailed explanations of what each machine/person was doing. We were then talked through the process of obtaining the genome sequence of our strains, from raw genomic DNA to the assembled sequence. <br/><br/> | ||
| + | |||
| + | <i>We would like to kindly thank two synthetic biologists, Dr. Wendy Smith, who spent her whole day enlightening us and Dr. Anil Wipat, who, we found out, was running an MSc Synthetic biology course.</i> | ||
| + | </p> | ||
| + | </div><!-- /article-text --> | ||
| + | </div><!-- /article --> | ||
| + | <div class="article" id="w05-26-07"> | ||
| + | <div class="article-title"> | ||
| + | <p><a name="week05-26-07">Week 5, 26.07.2012, Thursday</a></p> | ||
| + | </div> | ||
| + | <div class="article-details"> | ||
| + | <p><Posted on 1/8/2012></p> | ||
| + | </div> | ||
| + | <div class="article-text"> | ||
| + | <p> | ||
| + | <b>1) Herculase PCR for Shewanella </b><br/> | ||
| + | Herculase PCR was performed for potential Shewanella oneidensis strain using 3 sets of primers: cymA, mtrA and mtrCAB. Gel analysis indicated no results.<br/><br/> | ||
| + | <b>2) Citrobacter and E. coli growth on various sugars II and III – results</b><br/> | ||
| + | The results of these experiments can be seen in the pictures below. | ||
| + | <img style="width:612px;" src="http://i1056.photobucket.com/albums/t366/edigem12/260712/SugarsII.jpg" /><br/><br/> | ||
| + | <img style="width:612px;" src="http://i1056.photobucket.com/albums/t366/edigem12/010812/SugarsIII.jpg" /><br/> | ||
| + | From these results, it can be seen that Cf SBS197 grows less well on lactose, sucrose and citrate but both strains grow equally well on glycerol and glucose. The E. coli + sucrose hydrolase cells grew well on sucrose even without there being any arsenic (the inducer of the sucrose hydrolase gene) on the plate. For some reason, all bacteria grew weakly on the lactose plates, this might mean that our lactose stock might not be good. | ||
| + | |||
| + | |||
| + | </p> | ||
| + | </div><!-- /article-text --> | ||
| + | </div><!-- /article --> | ||
| + | <div class="article" id="w05-27-07"> | ||
| + | <div class="article-title"> | ||
| + | <p><a name="week05-27-07">Week 5, 27.07.2012, Friday</a></p> | ||
| + | </div> | ||
| + | <div class="article-details"> | ||
| + | <p><!-- Posted on ... --></p> | ||
| + | </div> | ||
| + | <div class="article-text"> | ||
| + | <p> | ||
| + | <b> Shewanella PCR </b> | ||
| + | Multiple PCRs were prepared:</br> | ||
| + | Kod PCR for 16S ribosomal DNA for potential Shewanella strain (strains was later identified as different bacteria)<br/> | ||
| + | Taq PCR for 3 potential Shewanella strains using mtrA primers (one of the strains showed positive result at expected 1 kb)<br/><br/> | ||
| + | <b> Half-cell setup</b> | ||
| + | Half-cell was autoclaved, filled with liquid LB medium and inocluated with potential Shewanella strain. The Half-cell was sealed and left for incubation over the weekend. The initial voltage readout was 0.4 mV. | ||
| + | </p> | ||
| + | </div><!-- /article-text --> | ||
| + | </div><!-- /article --> | ||
| + | </div><!-- /page-content --> | ||
| + | </div><!-- /page-middle --> | ||
</html> | </html> | ||
Latest revision as of 09:13, 8 August 2012
1) ccm transformants subculture
ccm transformed cells that lost RFP were subcultured.
Citrobacter and E. coli growth on various sugars III
A final test involved adding the various sugars to the M9 medium in the bottle as opposed to either adding it to the plate before pouring the agar or adding them into the well on the plate. 5x100ml M9 medium bottles were prepared as before and autoclaved. The sugars (1ml) and thiamine hydrochloride (3.4 ml) were added to the bottles prior to the agar getting poured, with two bottles having no sugar added to them. After the agar had set, the plates were inoculated as before. One of the no sugar plates had solid citrate added to its middle, as before with the solid glucose and sucrose, to test the growth of Cf on this medium that gives it its name.
1) Liquid cultures preparation
subcultures of ccm transformed cells were used to inoculate 3ml LB; samples were left to grow overnight
2) Primers preparation
Stock solution (500 pmol/ul, solution in EB) and working solution (10 pmol/ul 1 in 50 silution of stock solution in water) of primers were prepared for nfsl, cymA, mtrA and mtrCAB gene primers.
3) Kod PCR of Shewanella
Kod Pcr was performed for potential Shewanella oneidensis strain using 3 sets of primers: cymA, mtrA and mtrCAB. Gel analysis indicated no results.
4)Newcastle!
Eugene, Oscar and Réka travelled long and far to Newcastle to have the nice people there sequence our two Cf strains with their new Ion torrent sequencing suite. We also got the grand tour of their Centre for Bacterial Cell Biology, located in the new Baddiley-Clark Building, with detailed explanations of what each machine/person was doing. We were then talked through the process of obtaining the genome sequence of our strains, from raw genomic DNA to the assembled sequence.
We would like to kindly thank two synthetic biologists, Dr. Wendy Smith, who spent her whole day enlightening us and Dr. Anil Wipat, who, we found out, was running an MSc Synthetic biology course.
1) Herculase PCR for Shewanella
Herculase PCR was performed for potential Shewanella oneidensis strain using 3 sets of primers: cymA, mtrA and mtrCAB. Gel analysis indicated no results.
2) Citrobacter and E. coli growth on various sugars II and III – results
The results of these experiments can be seen in the pictures below.
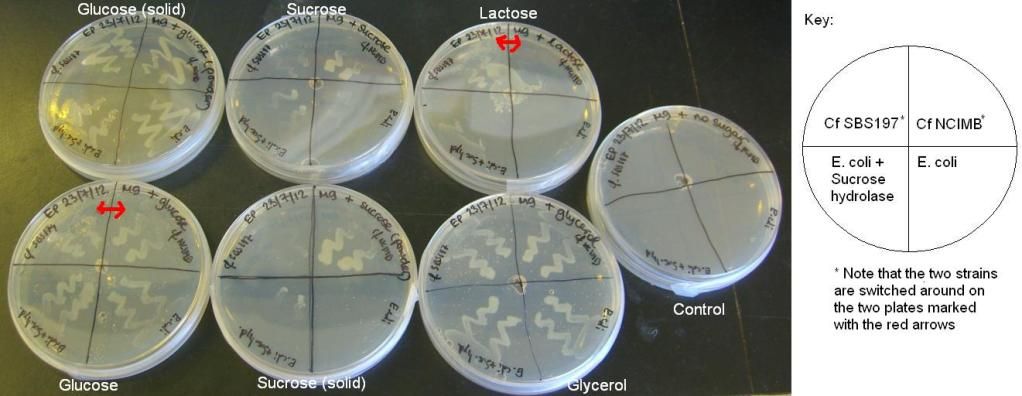
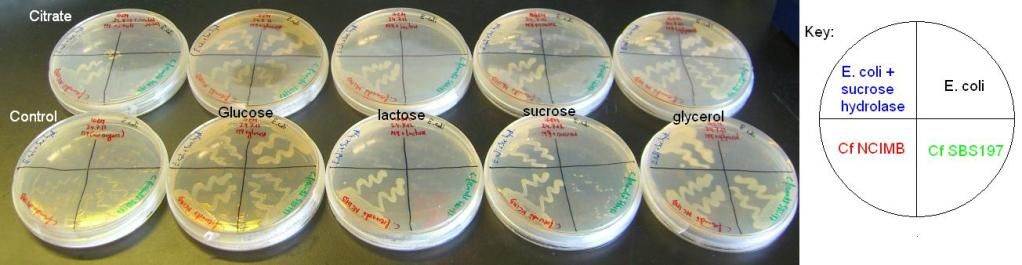
From these results, it can be seen that Cf SBS197 grows less well on lactose, sucrose and citrate but both strains grow equally well on glycerol and glucose. The E. coli + sucrose hydrolase cells grew well on sucrose even without there being any arsenic (the inducer of the sucrose hydrolase gene) on the plate. For some reason, all bacteria grew weakly on the lactose plates, this might mean that our lactose stock might not be good.
Shewanella PCR
Multiple PCRs were prepared:
Kod PCR for 16S ribosomal DNA for potential Shewanella strain (strains was later identified as different bacteria)
Taq PCR for 3 potential Shewanella strains using mtrA primers (one of the strains showed positive result at expected 1 kb)
Half-cell setup
Half-cell was autoclaved, filled with liquid LB medium and inocluated with potential Shewanella strain. The Half-cell was sealed and left for incubation over the weekend. The initial voltage readout was 0.4 mV.
 "
"







