Team:ZJU-China/models.htm
From 2012.igem.org
(Difference between revisions)
| (2 intermediate revisions not shown) | |||
| Line 140: | Line 140: | ||
align: justify; | align: justify; | ||
} | } | ||
| - | + | b.orange{ | |
| + | color: #FF8000; | ||
| + | font-style:italic; | ||
| + | } | ||
.projectNav a { | .projectNav a { | ||
font-size: 18px; | font-size: 18px; | ||
| Line 311: | Line 314: | ||
<div class="projectNav"> | <div class="projectNav"> | ||
| - | <p align="justify">In this year’s project, we try to make reasonable design of riboscaffolds by ourselves. Due to the current experimental conditions, we could not know what exactly happens to our riboscaffolds in the cell. Its folding process, binding with proteins and ligand, and the allosteric transition are of interests, since these are considerations for a reasonable design. Luckily, we have simulation tools for molecular modeling which can provide hints about what may happen theoretically in the intracellular environment. Although molecular modeling is often considered as professional work in which knowledge of different disciplines such as biology, physics, chemistry, mathematics and computer science as well as previous experience all contribute to good results, our team is willing to take a step in this field and get preliminary results.</p> | + | <p align="justify">In this year’s project, we try to make <b class="orange">reasonable design</b> of riboscaffolds by ourselves. Due to the current experimental conditions, we could not know what exactly happens to our riboscaffolds in the cell. Its folding process, binding with proteins and ligand, and the allosteric transition are of interests, since these are considerations for a reasonable design. Luckily, we have <b class="orange">simulation tools</b> for molecular modeling which can provide hints about <b class="orange">what may happen theoretically in the intracellular environment</b>. Although molecular modeling is often considered as professional work in which knowledge of different disciplines such as biology, physics, chemistry, mathematics and computer science as well as previous experience all contribute to good results, our team is willing to take a step in this field and get preliminary results.</p> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<div class="projectNavFloat"> | <div class="projectNavFloat"> | ||
| - | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_1.htm">1. RNA folding and 3D Structures</a> | + | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_1.htm" style="text-decoration:none">1. RNA folding and 3D Structures</a> |
<br> | <br> | ||
| - | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_2.htm">2. Docking</a> | + | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_2.htm" style="text-decoration:none">2. Docking</a> |
</div><!-- end .projectNavFloat --> | </div><!-- end .projectNavFloat --> | ||
<div class="projectNavFloat"> | <div class="projectNavFloat"> | ||
| - | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_3.htm">3. Molecular Dynamics</a> | + | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_3.htm" style="text-decoration:none">3. Molecular Dynamics</a> |
<br> | <br> | ||
| - | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_4.htm">4. References</a> | + | <a target="brainFrame" href="https://2012.igem.org/Team:ZJU-China/model_s1_4.htm" style="text-decoration:none">4. References</a> |
</div><!-- end .projectNavFloat --> | </div><!-- end .projectNavFloat --> | ||
| Line 342: | Line 345: | ||
<div class="projectNavFloat"> | <div class="projectNavFloat"> | ||
| - | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_1.htm">1. Preparation</a> | + | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_1.htm" style="text-decoration:none">1. Preparation</a> |
<br> | <br> | ||
| - | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_2.htm">2. A small scale simulation</a> | + | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_2.htm" style="text-decoration:none">2. A small scale simulation</a> |
</div><!-- end .projectNavFloat --> | </div><!-- end .projectNavFloat --> | ||
<div class="projectNavFloat"> | <div class="projectNavFloat"> | ||
| - | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_3.htm">3. A large scale simulation</a> | + | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_3.htm" style="text-decoration:none">3. A large scale simulation</a> |
<br> | <br> | ||
| - | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_4.htm">4. Additional effects</a> | + | <a target="brainFramei" href="https://2012.igem.org/Team:ZJU-China/model_s2_4.htm" style="text-decoration:none">4. Additional effects</a> |
</div><!-- end .projectNavFloat --> | </div><!-- end .projectNavFloat --> | ||
| Line 429: | Line 432: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
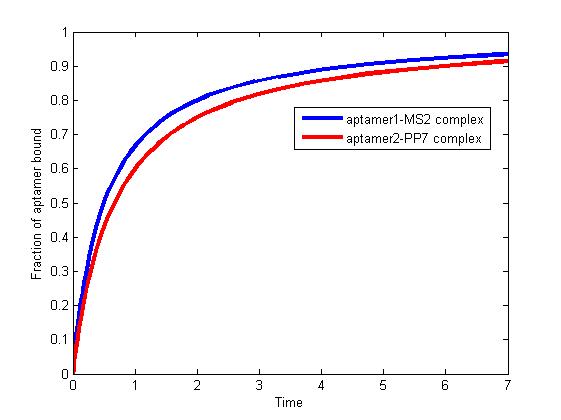
<p align="justify">The results are shown as follows:</p> | <p align="justify">The results are shown as follows:</p> | ||
| - | <div><img src="http://www.jiajunlu.com/igem/zju_model3_1.jpg"></div> | + | <div class="floatC"><img src="http://www.jiajunlu.com/igem/zju_model3_1.jpg"></div> |
<p class="fig" align="justify"><b>Fig 1.</b> The fraction of aptamer1-MS2 complex and the fraction of aptamer2-PP7 complex are increasing and finally reach equilibrium. </p> | <p class="fig" align="justify"><b>Fig 1.</b> The fraction of aptamer1-MS2 complex and the fraction of aptamer2-PP7 complex are increasing and finally reach equilibrium. </p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
Latest revision as of 03:14, 27 October 2012

 "
"