Team:Tokyo Tech
From 2012.igem.org
(→Ⅰ-4 Modeling) |
(→Ⅰ-2 Positive Feedback of cell-cell communication) |
||
| (201 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
=Project overview= | =Project overview= | ||
[[File:tokyotechprojectoverview.png|250px|right]] | [[File:tokyotechprojectoverview.png|250px|right]] | ||
| - | A love story contains several processes. Two people fall in love and their love burning wildly. However, no forever exists in the world, in most occasions, love will eventually burn to only a pile of ashes of the last remaining wind drift away. In our project, we have recreated the story of "Romeo and Juliet" by Shakespeare vividly by two kinds of Escherichia coli [[#Ⅰ cell-cell communication|cell-cell communication]]. We aim to generate a circuit involving regulatory mechanism of positive feedback rather than commonly-used negative feedback to control the fate of E.coli by signaling between two types of E.coli. Besides, Rose represents love. We are the first iGEM group ever to synthesize [[# | + | A love story contains several processes. Two people fall in love and their love burning wildly. However, no forever exists in the world, in most occasions, love will eventually burn to only a pile of ashes of the last remaining wind drift away. In our project, we have recreated the story of "Romeo and Juliet" by Shakespeare vividly by two kinds of <I>Escherichia coli</I> [[#Ⅰ cell-cell communication|cell-cell communication]]. We aim to generate a circuit involving regulatory mechanism of positive feedback rather than commonly-used negative feedback to control the fate of <I>E.coli</I> by signaling between two types of <I>E.coli</I>. Besides, Rose represents love. We are the first iGEM group ever to synthesize [[#.E2.85.A1__P.283HB.29_Production|P(3HB) ]] (a kind of bio-plastics) by using our new biobrick part, representing rose . |
</div class="whitebox"> | </div class="whitebox"> | ||
</div class="whitebox"> | </div class="whitebox"> | ||
| Line 18: | Line 18: | ||
=Ⅰ cell-cell communication= | =Ⅰ cell-cell communication= | ||
| - | "Romeo and Juliet" is the | + | "Romeo and Juliet" is the drama by dramatist William Shakespeare of England. The stage is city Verona, Italy in the 14th century. Romeo met Juliet. Two people fell in love instantly. In this project, we will recreate the love story of "Romeo and Juliet". |
| - | + | ||
| - | |||
| - | |||
| - | [[ | + | [[https://2012.igem.org/Team:Tokyo_Tech/Project Detailed descriptions for Cell-cell communication]] |
| + | ==Ⅰ-1 story== | ||
| + | We make our cute <I>E.coli</I> play “Romeo and Juliet” which is one of Shakespeare’s most famous plays. In this project, we define the signal that <I>E.coli</I> produce as the romantic feeling of Romeo and Juliet. In this project, we will recreate the love story of "Romeo and Juliet", by using "Cell-cell communication" | ||
The story that we reproduce is divided into four scenes. | The story that we reproduce is divided into four scenes. | ||
| - | '''( | + | Click on the Scene. |
| + | <html> | ||
| + | <script> | ||
| + | function show(k) | ||
| + | { | ||
| + | elem = document.getElementById('GNN'); | ||
| + | elem.style.display = 'none'; | ||
| + | if(k==1){ | ||
| + | elem.style.display = 'block'; | ||
| + | } | ||
| + | elem = document.getElementById('TNN'); | ||
| + | elem.style.display = 'none'; | ||
| + | if(k==2){ | ||
| + | elem.style.display = 'block'; | ||
| + | } | ||
| + | elem = document.getElementById('CNN'); | ||
| + | elem.style.display = 'none'; | ||
| + | if(k==3){ | ||
| + | elem.style.display = 'block'; | ||
| + | } | ||
| + | elem = document.getElementById('ANN'); | ||
| + | elem.style.display = 'none'; | ||
| + | if(k==4){ | ||
| + | elem.style.display = 'block'; | ||
| + | } | ||
| + | } | ||
| - | + | </script> | |
| - | + | <div id="marginbox"> | |
| - | + | <table align=center> | |
| - | < | + | <td><span id="student" onclick="show(1)">Scene1<br><IMG src="https://static.igem.org/mediawiki/2012/9/92/Tokyotechhhstory1.png" width="200px" border=0 |
| - | + | onMouseOver=this.src="https://static.igem.org/mediawiki/2012/2/2b/Tokyotechstorywaku1.png" width="10px" | |
| + | onMouseOut=this.src="https://static.igem.org/mediawiki/2012/9/92/Tokyotechhhstory1.png" width="200px"> | ||
| + | |||
| + | |||
| + | </br></span></td> | ||
| + | <td><span id="student" onclick="show(2)">Scene2<br><IMG src="https://static.igem.org/mediawiki/2012/2/21/Tokyotechhhstory2.png" width="200px" border=0 | ||
| + | onMouseOver=this.src="https://static.igem.org/mediawiki/2012/1/19/Tokyotechstorywaku2.png" width="200px" | ||
| + | onMouseOut=this.src="https://static.igem.org/mediawiki/2012/2/21/Tokyotechhhstory2.png" width="200px"> | ||
| + | |||
| + | </br></span></td> | ||
| + | <td><span id="student" onclick="show(3)">Scene3<br><IMG src="https://static.igem.org/mediawiki/2012/4/4e/Tokyotechhhstory3.png" width="200px" border=0 | ||
| + | onMouseOver=this.src="https://static.igem.org/mediawiki/2012/f/f9/Tokyotechstorywaku3.png" width="200px" | ||
| + | onMouseOut=this.src="https://static.igem.org/mediawiki/2012/4/4e/Tokyotechhhstory3.png" width="200px"> | ||
| + | |||
| + | </br></span></td> | ||
| + | <td><span id="student" onclick="show(4)">Scene4<br><IMG src="https://static.igem.org/mediawiki/2012/a/a6/Tokyotechhhstory4.png" width="200px" border=0 | ||
| + | onMouseOver=this.src="https://static.igem.org/mediawiki/2012/4/44/Tokyotechstoruwaku4.png" width="200px" | ||
| + | onMouseOut=this.src="https://static.igem.org/mediawiki/2012/a/a6/Tokyotechhhstory4.png" width="200px"> | ||
| + | |||
| + | </br></span></td> | ||
| + | </align> | ||
| + | </table> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | </html> | |
| - | + | <div id="GNN" style="width:900px"><div class="greybox"> | |
| + | [[File:tokyotechhstory1.png|300px|left]] | ||
| + | <p>'''(Scene 1)''' Romeo meets and falls in love with Juliet. Once the love between two people stimulates each other, they become deeply attached and cannot live without each other.<br><br><br>First, to represent the condition that their love stimulates each other in “Scene1 Fall in love”, we designed a positive feedback system in which the production of a signal activates the production of the other signal. [[https://2012.igem.org/Team:Tokyo_Tech/Project#Assays_for_Positive_feedback_system Detailed descriptions for Positive feedback system]] | ||
| + | </p> | ||
| + | <br><br> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div id="TNN" style="display:none"><div class="greenbox"> | ||
| + | [[File:tokyotechhstory2.png|300px|left]] | ||
| - | + | <p>'''(Scene 2)''' However, Juliet knows that their love will not be accepted by society because of family feud. To keep their relationship, Juliet plans to pretend to be dead. She takes a sleeping potion that makes her fall into a deathlike sleep. <br><br><br>Second, we applied a 3OC6HSL-dependent band detect system to represent “Scene2 Juliet’s deathlike sleep” by the stop of 3OC12HSL production in Juliet cell. When the concentration of 3OC6HSL reaches higher level by the positive feedback, the concentration of TetR is enough level to repress the expression of LasI, As a result, the production of 3OC12HSL is stopped though cell Juliet is alive. [[https://2012.igem.org/Team:Tokyo_Tech/Project#Band_detect_system Detailed descriptions for Band detect system]] | |
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div id="CNN" style="display:none"><div class="bluebox"> | ||
| + | [[File:tokyotechhstory3.png|300px|left]] | ||
| - | Finally, to realize “Scene4 Juliet’s suicide” in response to Romeo’s suicide, we also designed communication-inverter dependent suicide system in Juliet cell. After the death of Romeo cell, supply of 3OC6HSL is stopped, and the expression of LacI is stopped. As a result, the lysis gene is expressed and Juliet cell dies. | + | <p>'''(Scene 3)''' Romeo has heard of Juliet’s death without knowing the fact that Juliet is alive. Romeo decides to commit suicide by taking poison in response to Juliet’s “deathlike sleep”. <br><br><br>Third, to realize “Scene3 Romeo’s suicide” after Juliet fell into the deathlike sleep, we designed communication-inverter dependent suicide system in Romeo cell. When Juliet cell is in deathlike sleep, supply of 3OC12HSL is stopped though Juliet cell is alive. In the absence of 3OC12HSL, the lysis gene is expressed and Romeo cell dies.[[https://2012.igem.org/Team:Tokyo_Tech/Project#communication-inverter_dependent_suicide_system Detailed descriptions for communication-inverter dependent suicide system]] |
| + | </p> | ||
| + | <br> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div id="ANN" style="display:none"><div class="redbox"> | ||
| + | [[File:tokyotechhstory4.png|300px|left]] | ||
| + | |||
| + | <p>'''(Scene 4)''' Juliet awakes to find Romeo dead beside her. She decides to commit suicide in response to Romeo’s suicide. She stabs herself with a dirk.<br><br><br>Finally, to realize “Scene4 Juliet’s suicide” in response to Romeo’s suicide, we also designed communication-inverter dependent suicide system in Juliet cell. After the death of Romeo cell, supply of 3OC6HSL is stopped, and the expression of LacI is stopped. As a result, the lysis gene is expressed and Juliet cell dies.[[https://2012.igem.org/Team:Tokyo_Tech/Project#communication-inverter_dependent_suicide_system Detailed descriptions for communication-inverter dependent suicide system]] | ||
| + | </p> | ||
| + | |||
| + | <br><br> | ||
| + | </div> | ||
| + | </div> | ||
==Ⅰ-2 Positive Feedback of cell-cell communication== | ==Ⅰ-2 Positive Feedback of cell-cell communication== | ||
| - | [[File: | + | [[File:positivefeedbackassay20tokyotech.png|800px|thumb|center|Fig1-2,[[https://2012.igem.org/Team:Tokyo_Tech/Project#Positive_feedback_assay.7ETime-dependent_change_assay.7E Time-dependent change assay]]]] |
| - | + | ||
| - | [[https://2012.igem.org/Team:Tokyo_Tech/Project | + | |
| - | + | To accomplish the positive feedback system in the cell-cell communication, we designed and constructed two types of genetically engineered <I>E.coli</I>, 3OC6HSL-dependent 3OC12HSL producer cell (Plux-LasI cell) and 3OC12HSL-dependent 3OC6HSL producer cell (Plas-LuxI cell). | |
| - | + | <br>For a trigger of the positive feedback system, we added the initial dose of 3OC6HSL (5 nM) to the co-cultures. Only when the two types of signal producer cells were cultured together and allowed to communicate with one another, signal content in the supernatant of the co-culture drastically increase as compared to other conditions. <br>As compared red solid line with blue dotted line in the condition i (both Plux-LasI cell and Plas-LuxI cell coexist), the fig shows that the fluorescence intensity of Las reporter increases at first (0-1h), and then that of Lux reporter starts to increase (1-2h). This result indicates that the 3OC12HSL production in Plux-LasI cell was activated by initially added 3OC6HSL, whereas the 3OC6HSL production in Plas-LuxI cell was not activated till 3OC12HSL production in Plux-LasI cell reached sufficient level. This behavior strongly suggests the appearance of the positive feedback. | |
| - | < | + | <br>In the process of the implementation, we also confirmed two modules, the 3OC6HSL-dependent 3OC12HSL production module and the 3OC12HSL-dependent 3OC6HSL production module, which constituted our positive feedback system. In these module, two new Biobrick parts, Plux-LasI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934022 BBa_K934022]) and Plas-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]), were characterized their functions. |
| - | <br><br> | + | [[https://2012.igem.org/Team:Tokyo_Tech/Project Detailed descriptions for Cell-cell communication]] |
| - | <br> | + | |
| - | + | ||
<div id="tokyotech" style=" font:bold ;left ; font-size: 20px; color: #000000; padding: 10px;"> | <div id="tokyotech" style=" font:bold ;left ; font-size: 20px; color: #000000; padding: 10px;"> | ||
Other experiments for basis of "Romeo & Juliet" | Other experiments for basis of "Romeo & Juliet" | ||
</div> | </div> | ||
| - | [[File:positivefeedbackassay18tokyotech.png| | + | |
| - | [[File:positivefeedbackassay19tokyotech.png|150px|thumb|left| | + | [[File:positivefeedbackassay18tokyotech.png|170px|thumb|left|Fig1-3,[[https://2012.igem.org/Team:Tokyo_Tech/Project#Construction_of_the_3OC6HSL-dependent_3OC12HSL_production_module Go to <br>"Construction_of<br>the_3OC6HSL-dependent<br>3OC12HSL_production"]]]] |
| - | [[File: | + | [[File:positivefeedbackassay19tokyotech.png|170px|thumb|left|Fig1-4,[[https://2012.igem.org/Team:Tokyo_Tech/Project#Construction_of_the_3OC12HSL-dependent_3OC6HSL_production_module Go to <br>"Construction_of<br>the_3OC12HSL-dependent<br>3OC6HSL_production"]]]] |
| + | [[File:positivefeedbackassay30tokyotech.png|150px|thumb|left|Fig1-5,[[https://2012.igem.org/Team:Tokyo_Tech/Project#Positive_feedback_assay_.7ECo-culture_assay.7E Go to <br>"Co-culture assay"]]]] | ||
| + | |||
| + | [[File:positivefeedbackassay80tokyotech.png|150px|thumb|left|Fig1-6,[[https://2012.igem.org/Team:Tokyo_Tech/Project#Band_detect_system Go to <br>"Band detect system"]]]] | ||
| + | |||
| + | |||
<br><br><br><br><br><br> | <br><br><br><br><br><br> | ||
| - | + | <br><br> | |
| - | <br><br><br> | + | <br><br> |
| - | <br> | + | <br><br> |
| - | ==Ⅰ- | + | ==Ⅰ-3 Modeling== |
| - | [[File:tokyotechModelingresult2.png|250px|thumb|right| | + | [[File:tokyotechModelingresult2.png|250px|thumb|right|Fig1-7, time-dependent change of the concentrations of the two signals.]] |
| - | [[File:tokyotechStory.png|200px|thumb|right| | + | [[File:tokyotechStory.png|200px|thumb|right|Fig1-8, the story of “Romeo and Juliet”]] |
'''Result: Whether our circuit can reproduce “Romeo and Juliet”''' | '''Result: Whether our circuit can reproduce “Romeo and Juliet”''' | ||
| - | To confirm the feasibility of the cell-cell communication system, we developed an ordinary differential equation model and simulated the system under typical experimental conditions. | + | To confirm the feasibility of the cell-cell communication system, we developed an ordinary differential equation model and simulated the system under typical experimental conditions. Fig1-7 shows the result of the simulation. As described below, the behavior of the signal concentration is consistent with the development of the “Romeo and Juliet” story. |
| - | '''In the | + | '''In the yellow area of Fig1-7''', the concentration of two signals increases. It represents Scene1. |
| - | '''In the green area of | + | '''In the green area of Fig1-7''', as the concentration of Romeo signals increases to some extent, the concentration of Juliet signals starts to decline. It represents Scene2. |
| - | '''In the blue area of | + | '''In the blue area of Fig1-7''', lysis gene is expressed in Romeo cell in response to decline of the concentration of Juliet signals, then the concentration of Romeo signals starts to decline. It represents Scene3. |
| - | '''In the pink area of | + | '''In the pink area of Fig1-7''', lysis gene is expressed in Juliet cell in response to the decline of the concentration of Romeo signals, then the concentration of Juliet signals decreases further. It represents Scene4. |
| + | |||
| + | [[https://2012.igem.org/Team:Tokyo_Tech/Project#Modeling Detailed descriptions for Modeling]] | ||
<br><br><br><br> | <br><br><br><br> | ||
| - | =Ⅱ | + | =Ⅱ P(3HB) Production= |
==Ⅱ-1 Story== | ==Ⅱ-1 Story== | ||
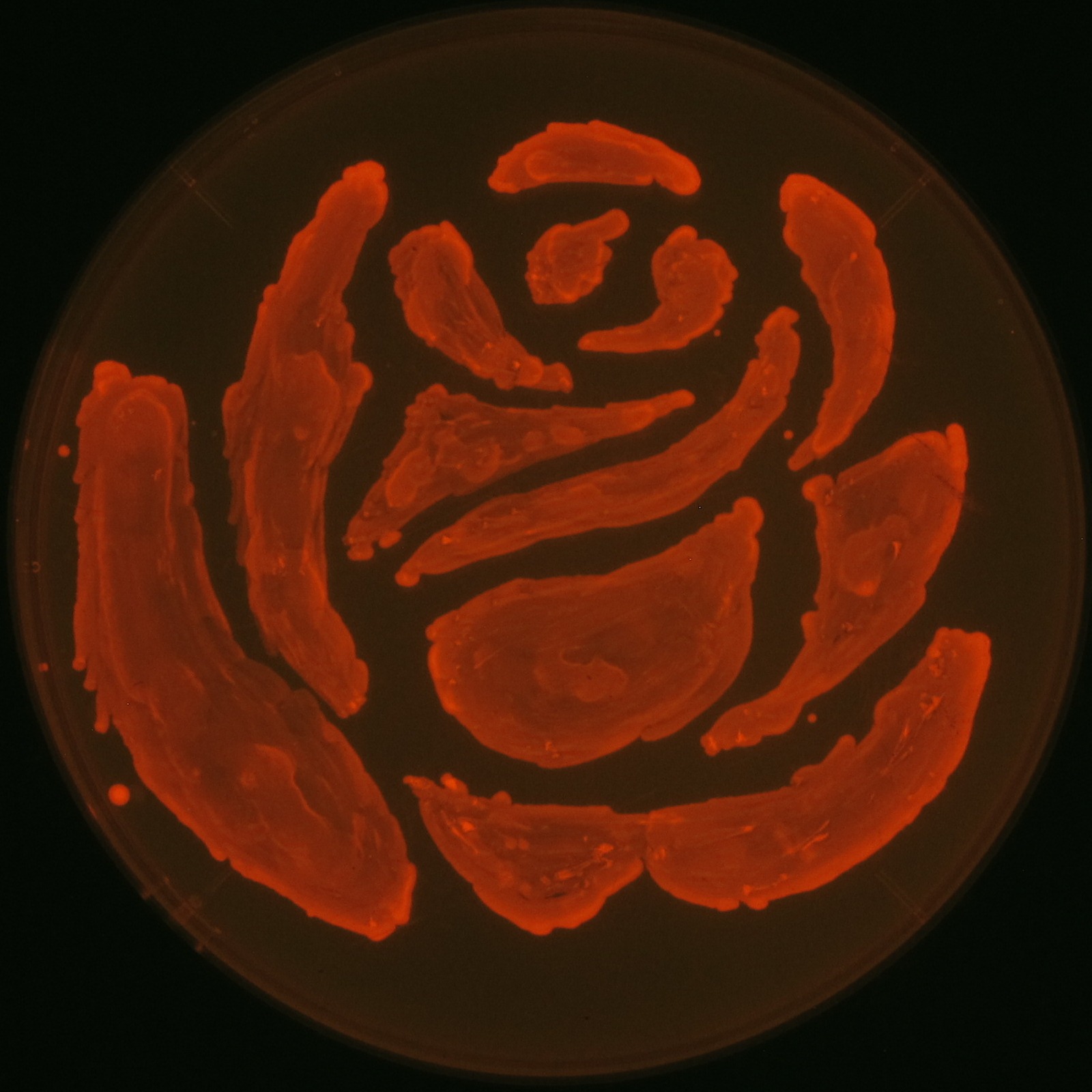
| - | [[File: | + | [[File:tokyotech PHA make rose.png|300px|thumb|right|fig1-9,Rose silhouette on the LB agar plate containing Nile red.]] |
| - | There is what of the famous scene of "Romeo and | + | |
| + | There is what of the famous scene of "Romeo and Juliet" | ||
<div style=" font:bold ;left ; font-size: 15px; color: #8B0000; padding: 0px;"> | <div style=" font:bold ;left ; font-size: 15px; color: #8B0000; padding: 0px;"> | ||
JULIET: O Romeo, Romeo! why are you Romeo? Deny your father and refuse your name;</div> | JULIET: O Romeo, Romeo! why are you Romeo? Deny your father and refuse your name;</div> | ||
| Line 103: | Line 176: | ||
JULIET: It's but your name that is my enemy; you are yourself, though not a Montague. O, be some other name! What's in a name? that which we call a rose By any other name would smell as sweet.</div> | JULIET: It's but your name that is my enemy; you are yourself, though not a Montague. O, be some other name! What's in a name? that which we call a rose By any other name would smell as sweet.</div> | ||
we will recreate the rose come out in the lines of the famous drama "Romeo and Juliet" | we will recreate the rose come out in the lines of the famous drama "Romeo and Juliet" | ||
| - | by the synthesis of | + | by the synthesis of P(3HB). |
| - | ==Ⅱ-2 | + | ==Ⅱ-2 P(3HB) production== |
| - | + | We made a new biobrick part and succeeded in synthesizing Polyhydroixyalkanoates(PHAs). This is the first Biobrick part to synthesize P(3HB), a kind of PHAs. | |
| - | We made a new biobrick part and succeeded synthesizing Polyhydroixyalkanoates(PHAs). This is the first Biobrick part to synthesize PHAs. | + | In our project, we also draw rose silhouette to produce the balcony scene of “Romeo and Juliet” by the synthesis of P(3HB).[[https://2012.igem.org/Team:Tokyo_Tech/Projects/PHAs/index.htm Detailed descriptions for P(3HB) production]] |
| - | In our project, we also draw rose silhouette to produce the balcony scene of “Romeo and Juliet” by the synthesis of | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div class="whitebox"> | </div class="whitebox"> | ||
</div class="whitebox"> | </div class="whitebox"> | ||
<div class="whitebox"> | <div class="whitebox"> | ||
| + | <div id="tokyotech" style=" font:bold ;left ; font-size: 15px; color: #000000; padding: 10px;"> | ||
| - | = | + | =Human Practice= |
| - | + | [[File:tokyotech human7.png|200px|right]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Taku Nakayama, and Mai Miura (members of Tokyo_Tech iGEM team) have participated in a science cafe as assistants for the event. The two members supported people who are not specialist in biology to plan an imaginary synthetic biology project, to be evaluated from public points of view, and to upgrade the project in accordance with the evaluations. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[https://2012.igem.org/Team:Tokyo_Tech/HumanPractice.htm Detailed descriptions for Human practice] | |
| - | + | ] | |
| - | + | <br> | |
| - | + | ||
Latest revision as of 03:10, 27 October 2012
Project overview
A love story contains several processes. Two people fall in love and their love burning wildly. However, no forever exists in the world, in most occasions, love will eventually burn to only a pile of ashes of the last remaining wind drift away. In our project, we have recreated the story of "Romeo and Juliet" by Shakespeare vividly by two kinds of Escherichia coli cell-cell communication. We aim to generate a circuit involving regulatory mechanism of positive feedback rather than commonly-used negative feedback to control the fate of E.coli by signaling between two types of E.coli. Besides, Rose represents love. We are the first iGEM group ever to synthesize P(3HB) (a kind of bio-plastics) by using our new biobrick part, representing rose .
Ⅰ cell-cell communication
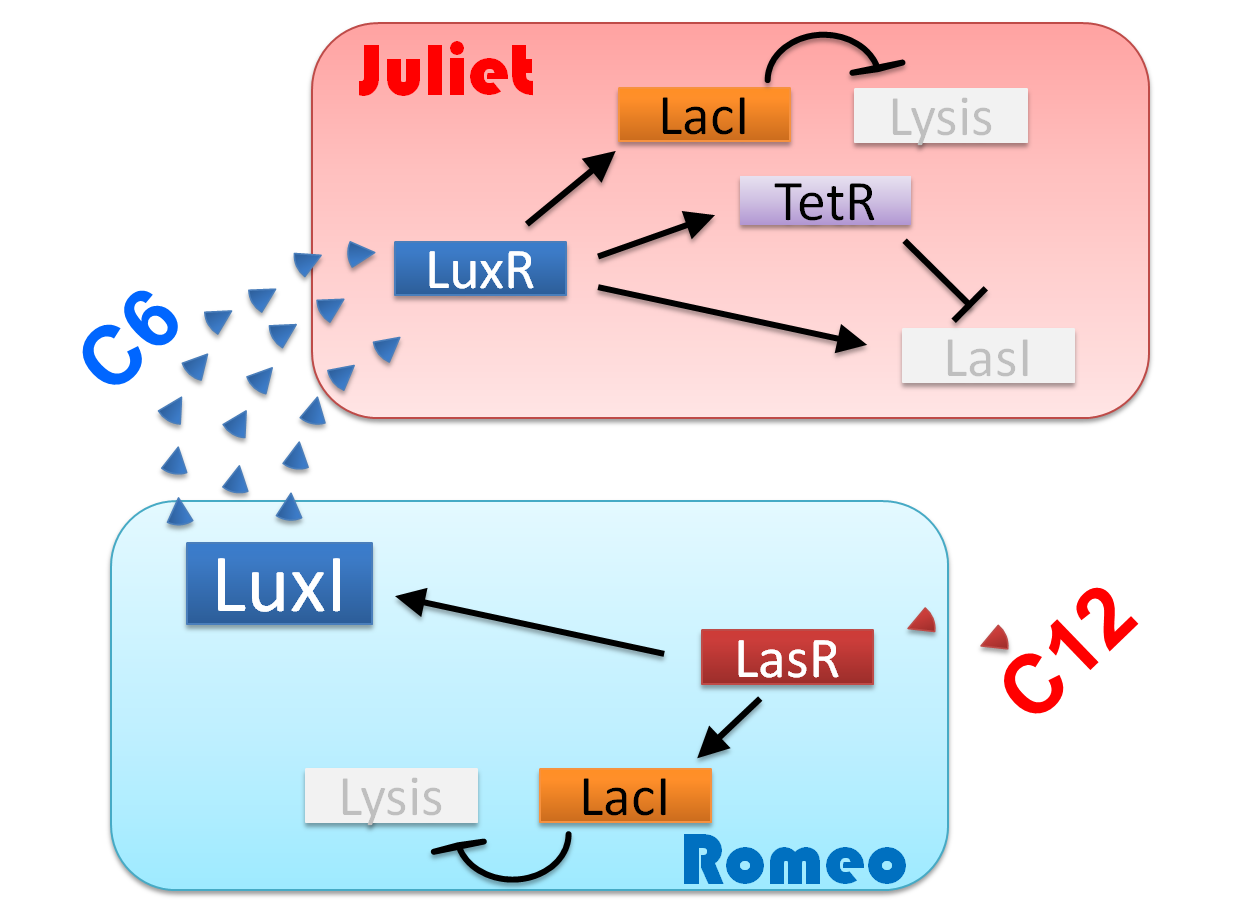
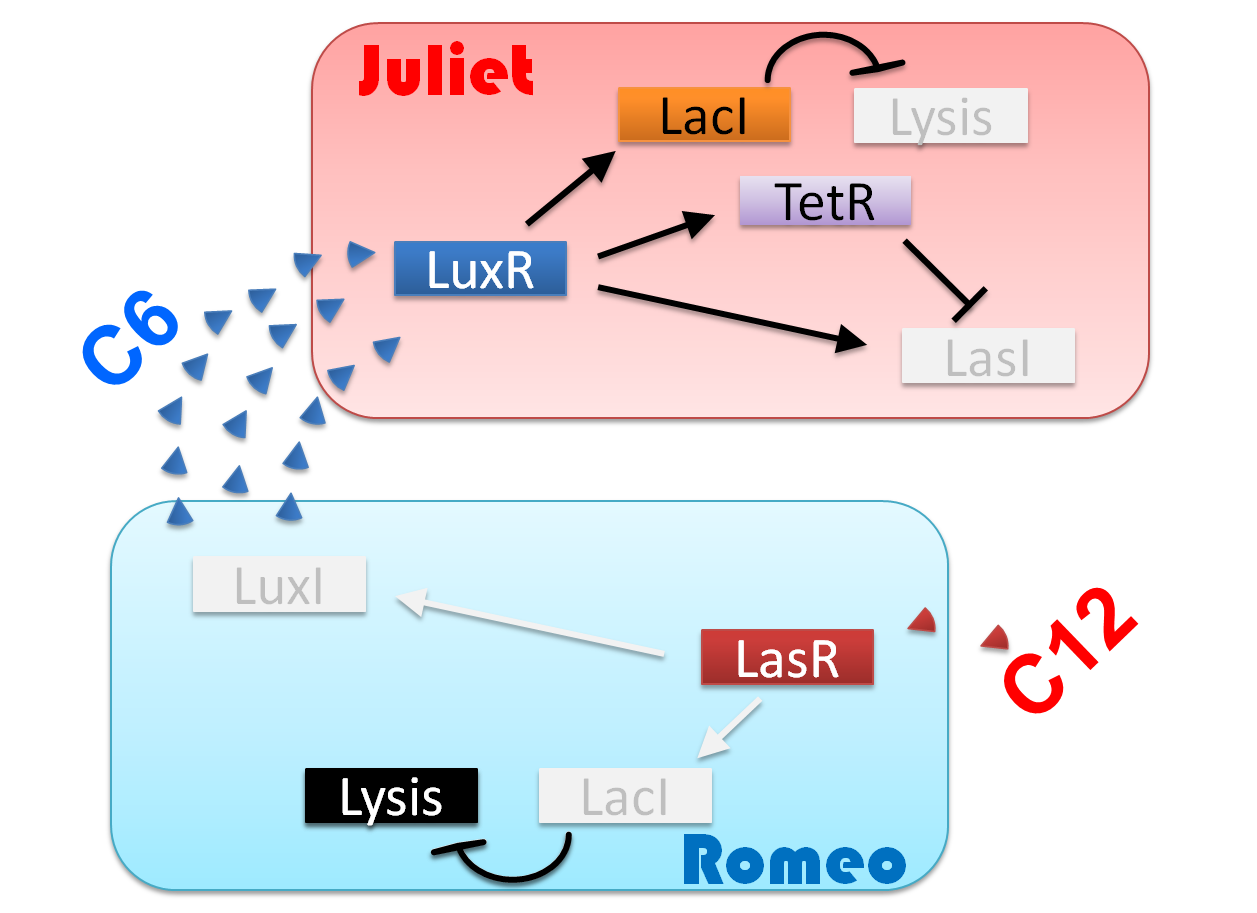
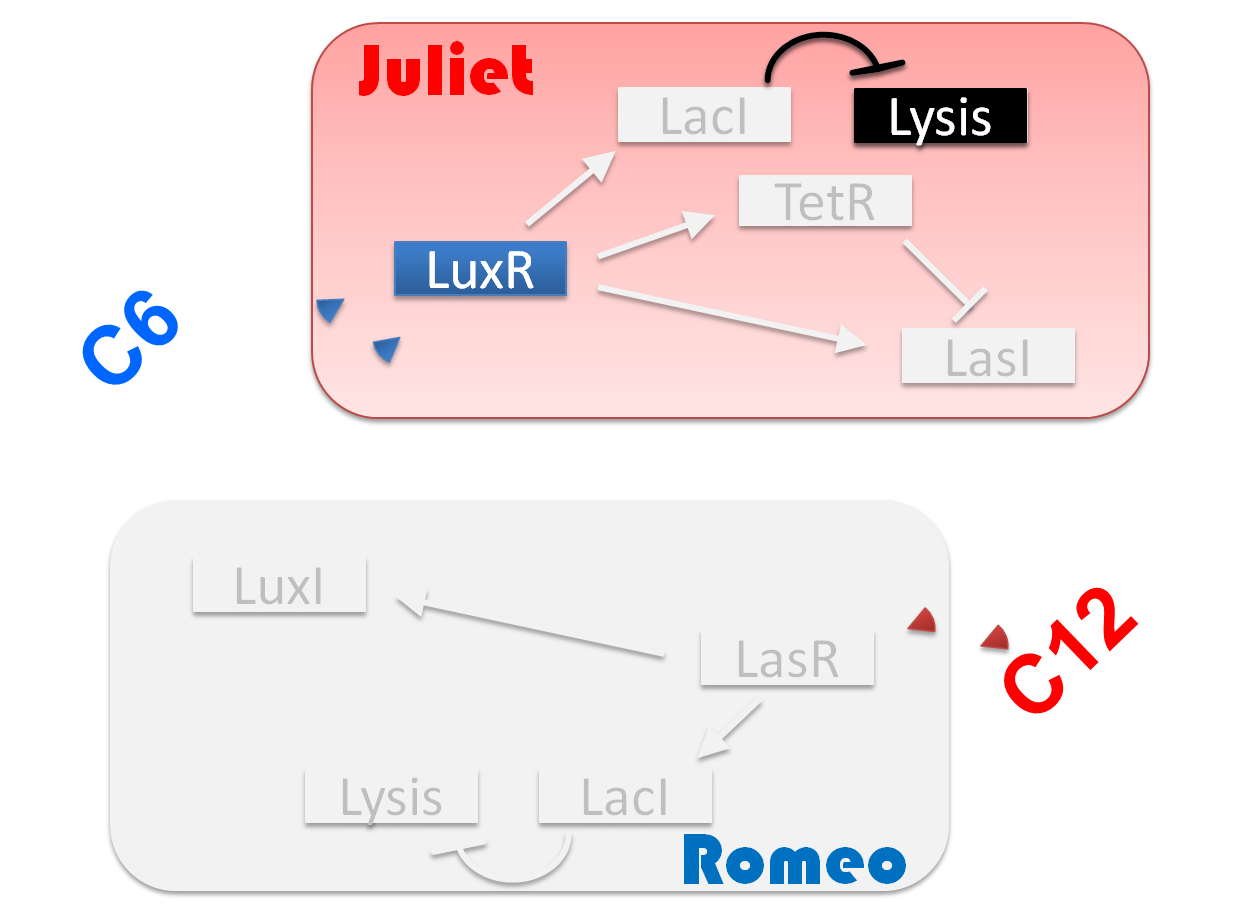
"Romeo and Juliet" is the drama by dramatist William Shakespeare of England. The stage is city Verona, Italy in the 14th century. Romeo met Juliet. Two people fell in love instantly. In this project, we will recreate the love story of "Romeo and Juliet".
[Detailed descriptions for Cell-cell communication]
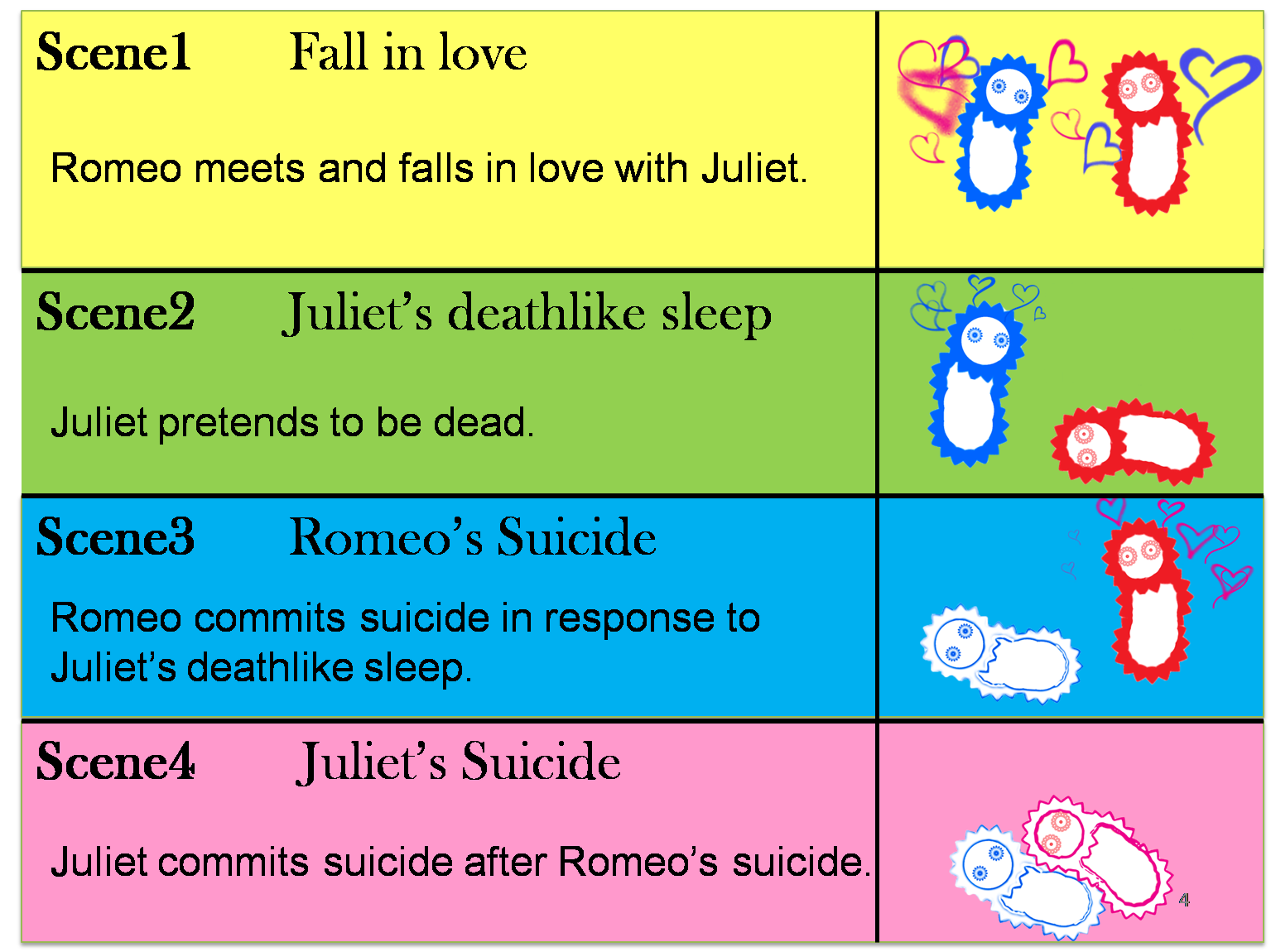
Ⅰ-1 story
We make our cute E.coli play “Romeo and Juliet” which is one of Shakespeare’s most famous plays. In this project, we define the signal that E.coli produce as the romantic feeling of Romeo and Juliet. In this project, we will recreate the love story of "Romeo and Juliet", by using "Cell-cell communication" The story that we reproduce is divided into four scenes.
Click on the Scene.
Scene1
|
Scene2
|
Scene3
|
Scene4
|
(Scene 1) Romeo meets and falls in love with Juliet. Once the love between two people stimulates each other, they become deeply attached and cannot live without each other.
First, to represent the condition that their love stimulates each other in “Scene1 Fall in love”, we designed a positive feedback system in which the production of a signal activates the production of the other signal. [Detailed descriptions for Positive feedback system]
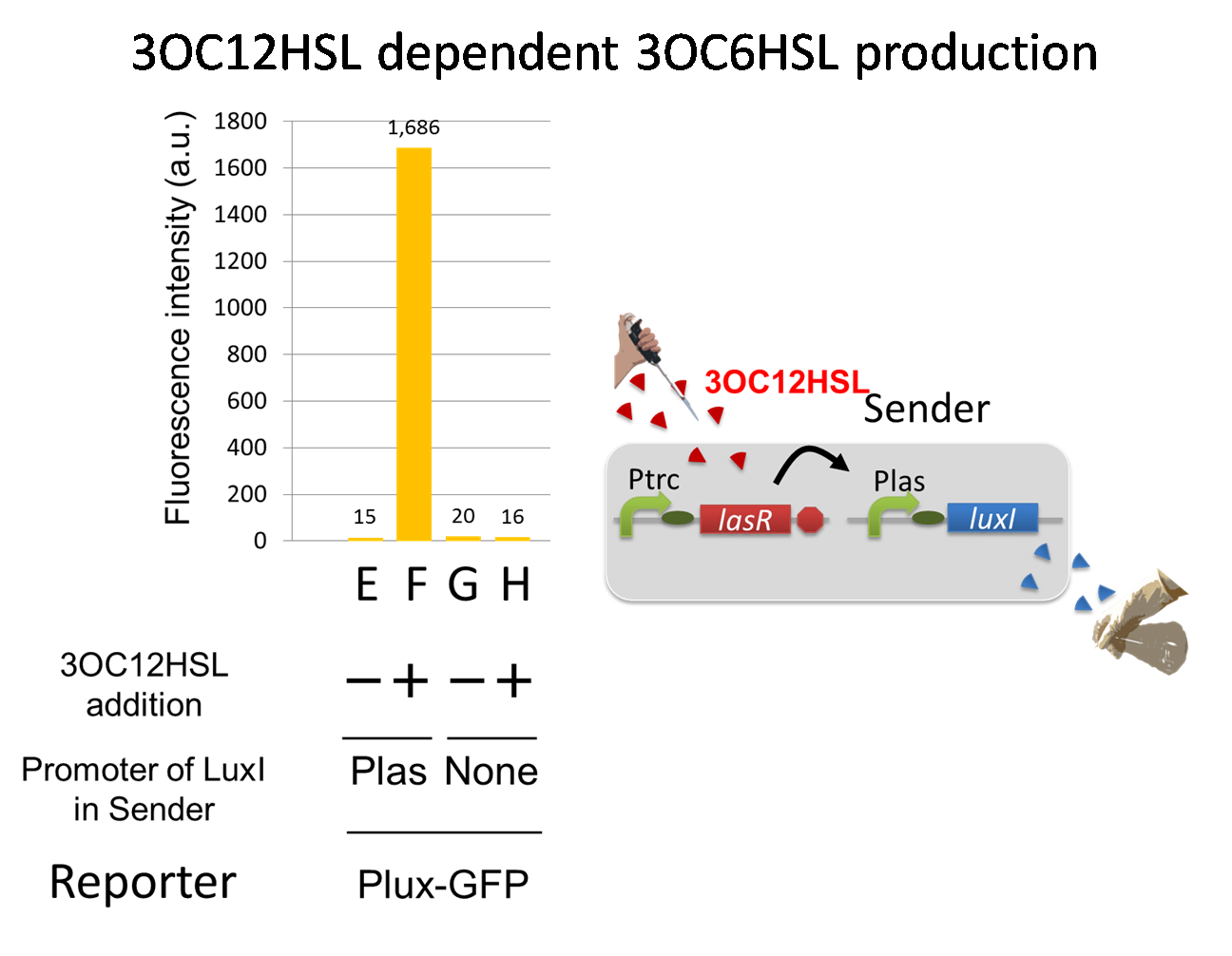
Ⅰ-2 Positive Feedback of cell-cell communication
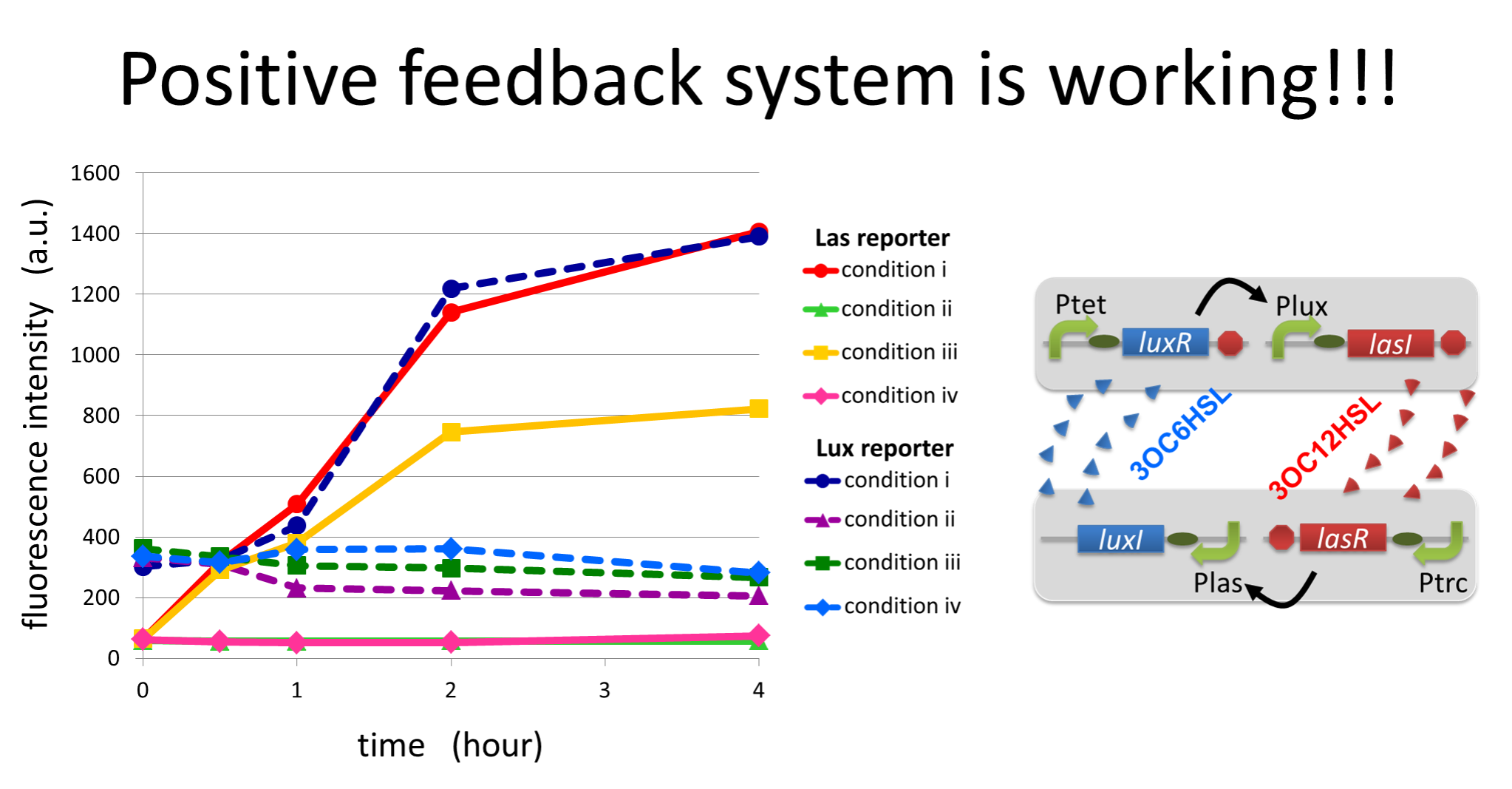
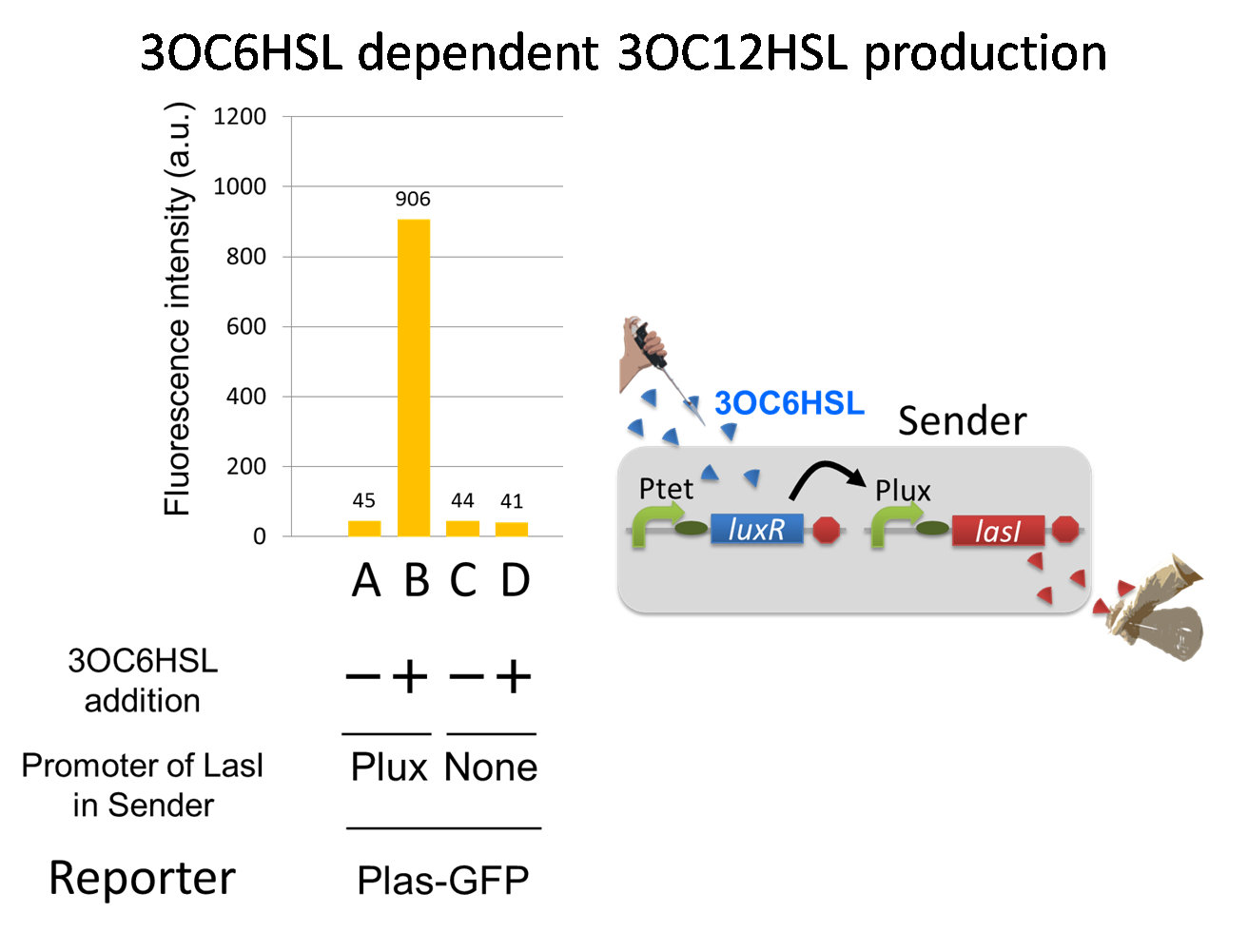
To accomplish the positive feedback system in the cell-cell communication, we designed and constructed two types of genetically engineered E.coli, 3OC6HSL-dependent 3OC12HSL producer cell (Plux-LasI cell) and 3OC12HSL-dependent 3OC6HSL producer cell (Plas-LuxI cell).
For a trigger of the positive feedback system, we added the initial dose of 3OC6HSL (5 nM) to the co-cultures. Only when the two types of signal producer cells were cultured together and allowed to communicate with one another, signal content in the supernatant of the co-culture drastically increase as compared to other conditions.
As compared red solid line with blue dotted line in the condition i (both Plux-LasI cell and Plas-LuxI cell coexist), the fig shows that the fluorescence intensity of Las reporter increases at first (0-1h), and then that of Lux reporter starts to increase (1-2h). This result indicates that the 3OC12HSL production in Plux-LasI cell was activated by initially added 3OC6HSL, whereas the 3OC6HSL production in Plas-LuxI cell was not activated till 3OC12HSL production in Plux-LasI cell reached sufficient level. This behavior strongly suggests the appearance of the positive feedback.
In the process of the implementation, we also confirmed two modules, the 3OC6HSL-dependent 3OC12HSL production module and the 3OC12HSL-dependent 3OC6HSL production module, which constituted our positive feedback system. In these module, two new Biobrick parts, Plux-LasI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934022 BBa_K934022]) and Plas-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]), were characterized their functions.
[Detailed descriptions for Cell-cell communication]
Other experiments for basis of "Romeo & Juliet"
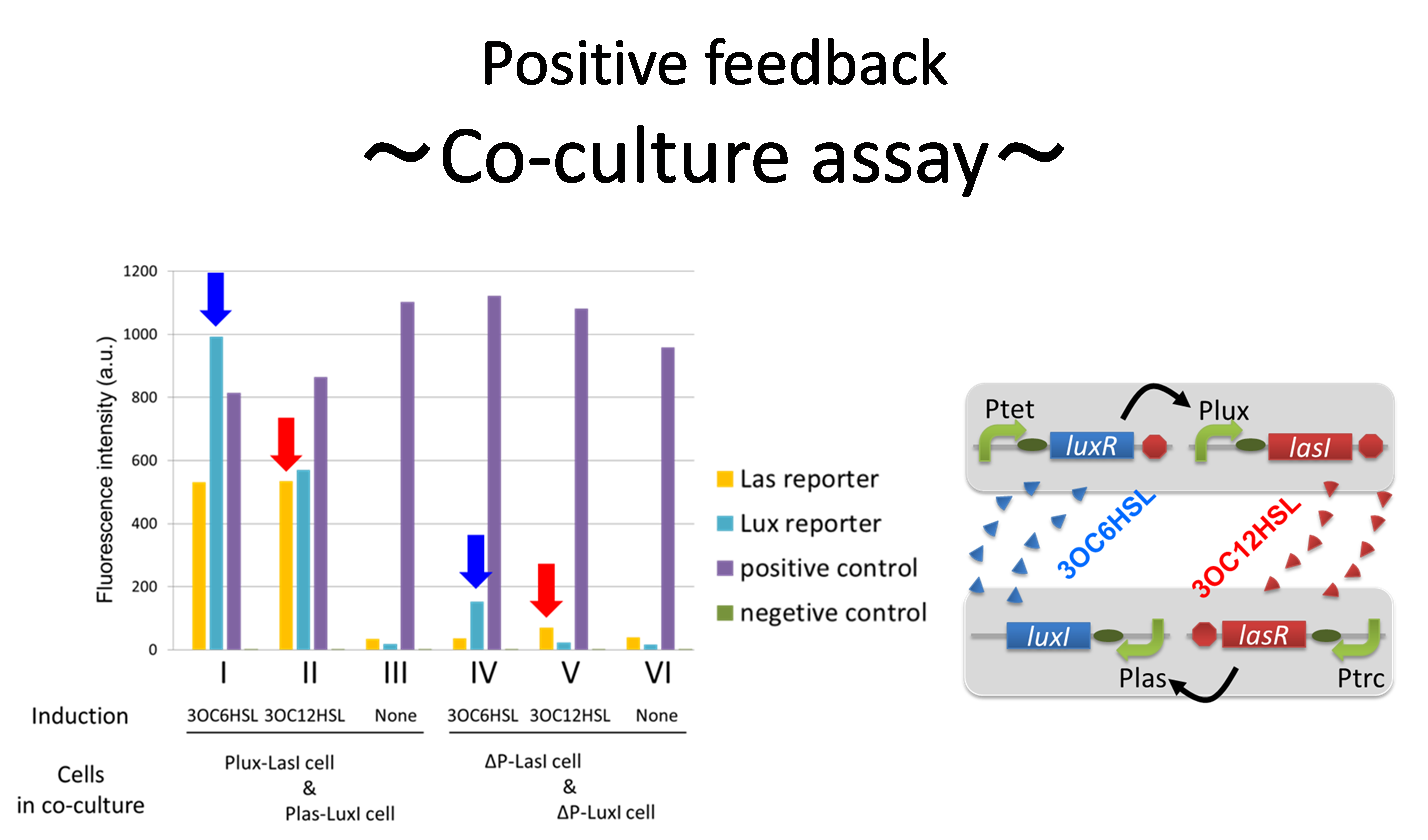
"Co-culture assay"]

"Band detect system"]
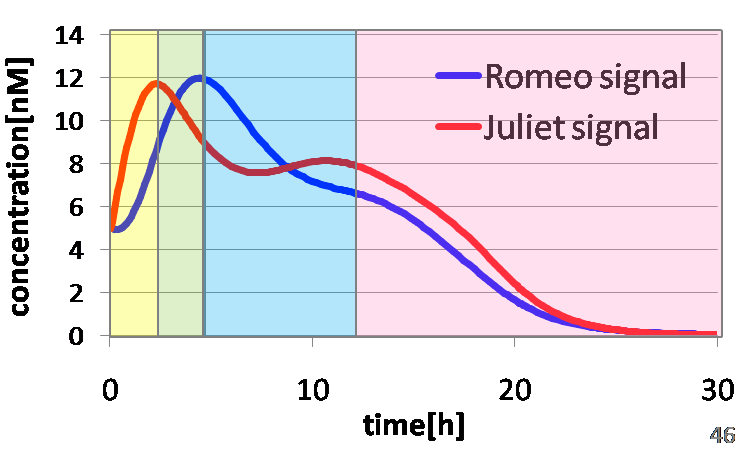
Ⅰ-3 Modeling
Result: Whether our circuit can reproduce “Romeo and Juliet”
To confirm the feasibility of the cell-cell communication system, we developed an ordinary differential equation model and simulated the system under typical experimental conditions. Fig1-7 shows the result of the simulation. As described below, the behavior of the signal concentration is consistent with the development of the “Romeo and Juliet” story.
In the yellow area of Fig1-7, the concentration of two signals increases. It represents Scene1.
In the green area of Fig1-7, as the concentration of Romeo signals increases to some extent, the concentration of Juliet signals starts to decline. It represents Scene2.
In the blue area of Fig1-7, lysis gene is expressed in Romeo cell in response to decline of the concentration of Juliet signals, then the concentration of Romeo signals starts to decline. It represents Scene3.
In the pink area of Fig1-7, lysis gene is expressed in Juliet cell in response to the decline of the concentration of Romeo signals, then the concentration of Juliet signals decreases further. It represents Scene4.
[Detailed descriptions for Modeling]
Ⅱ P(3HB) Production
Ⅱ-1 Story
There is what of the famous scene of "Romeo and Juliet"
we will recreate the rose come out in the lines of the famous drama "Romeo and Juliet" by the synthesis of P(3HB).
Ⅱ-2 P(3HB) production
We made a new biobrick part and succeeded in synthesizing Polyhydroixyalkanoates(PHAs). This is the first Biobrick part to synthesize P(3HB), a kind of PHAs. In our project, we also draw rose silhouette to produce the balcony scene of “Romeo and Juliet” by the synthesis of P(3HB).[Detailed descriptions for P(3HB) production]
Human Practice
Taku Nakayama, and Mai Miura (members of Tokyo_Tech iGEM team) have participated in a science cafe as assistants for the event. The two members supported people who are not specialist in biology to plan an imaginary synthetic biology project, to be evaluated from public points of view, and to upgrade the project in accordance with the evaluations.
[Detailed descriptions for Human practice ]
 "
"