Team:Bielefeld-Germany/Judging
From 2012.igem.org
(→How our BioBricks work) |
|||
| (135 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | <div id=page-title> | ||
| + | <span id=page-title-text> | ||
| + | Judging | ||
| + | </span> | ||
| + | </div> | ||
<style type="text/css"> | <style type="text/css"> | ||
| Line 27: | Line 32: | ||
</li> | </li> | ||
| + | |||
<li> | <li> | ||
| - | <a href="#3"> | + | <a href="#3">Collaboration<strong></strong> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</a> | </a> | ||
</li> | </li> | ||
| Line 42: | Line 44: | ||
<!-- tab panes --> | <!-- tab panes --> | ||
| - | <div id=" | + | <div id="judging_site"> |
| - | <div | + | <div id="anzeige"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <h1>Our BioBricks</h1> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </html> | |
| - | + | __NOTOC__ | |
| + | <groupparts>iGEM012 Bielefeld-Germany</groupparts> | ||
| + | <html> | ||
| + | |||
</div> | </div> | ||
| - | <div | + | <div id="anzeige"> |
| - | + | ||
| - | + | <h1>Datapage</h1> | |
| - | + | </html> | |
| - | + | ||
| - | + | == How our BioBricks work== | |
| - | + | ||
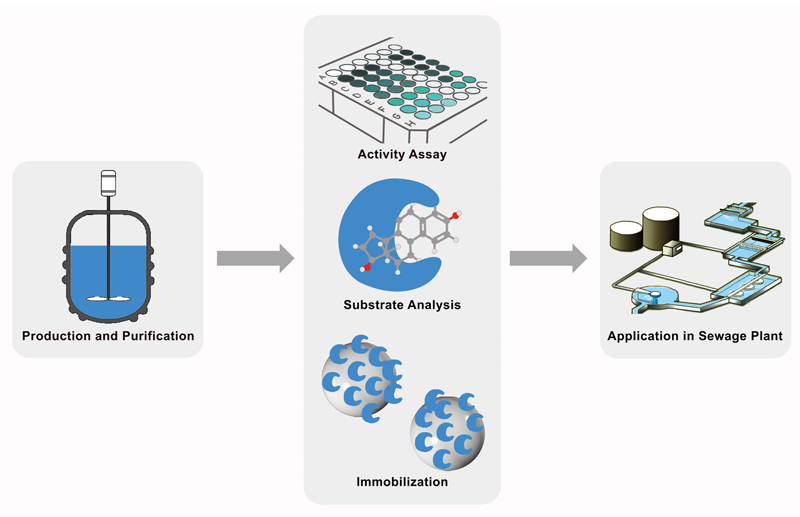
| - | + | [[File:Bielefeld2012 Overview.jpg|900px|thumb|center|'''Figure 1:''' iGEM Team Bielefeld is developing a biological filter using immobilized laccases, enzymes able to radicalize and break down a broad range of aromatic substances. For the production of laccases from different bacteria, fungi and plants, three expression systems were used: ''Escherichia coli'' KRX and Rosetta-Gami 2 and the yeast ''Pichia pastoris''. Immobilization is carried out either by using CPC-silica beads or by fusing the enzymes to cellulose binding domains. The concept could be extended to other toxic pollutants in drinking and wastewater, as well as to industrial applications in paper and textile industries or even for bioremediation of contaminated soil. | |
| - | + | ]] | |
| - | + | ||
| - | + | ==Data for our favorite new parts== | |
| - | + | # <partinfo>K863000</partinfo> - '''bpul (laccase from ''Bacillus pumilus'') with T7 promoter, RBS and His-tag''': This part is used to overexpress the laccase bpul for further purification followed by characterization of enzyme activity and substrate specificity. | |
| + | # <partinfo>K863005</partinfo> - '''ecol (laccase from ''E. coli'') with T7 promoter, RBS and His-tag''': This enzyme is overexpressed after induction and can be purified by His-tag. Subsequently the laccase can be characterized regarding to enzyme activity and substrate specificity. | ||
| + | # <partinfo>K863204</partinfo> - '''shuttle vector pECPP11JS for site-directed recombination of genes of interest in yeast''': With this part the production and secretion of a protein of interest like laccase is possible. | ||
| + | |||
| + | ==We have also characterized the following parts== | ||
| + | # <partinfo>BBa_K863012</partinfo> - '''tthl laccase (from ''T. thermophilus'') with constitutive promoter J23100, RBS and His-tag''': This part is used for the constitutive expression of tthl for further purification followed by characterization of enzyme activity and substrate specificity. | ||
| + | # <partinfo>BBa_K863022</partinfo> - '''bhal laccase (''from Bacillus halodurans'') with constitutive promoter J23100, RBS and His-tag''': This part is used for the constitutive expression of bhal for further purification followed by characterization of enzyme activity and substrate specificity. | ||
| + | # <partinfo>BBa_K863207</partinfo> - '''shuttle vector pECPP11JS for site-directed recombination of TVEL5 laccase in yeast:''' With this part the production and secretion of TVEL5 is possible. | ||
| + | <html> | ||
| + | |||
</div> | </div> | ||
| - | |||
| - | + | <div id="anzeige"> | |
| + | <h1>Collaborations</h1> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
| - | = | + | __NOTOC__ |
| + | <div style="text-align:justify;"> | ||
| - | == | + | == Collaboration with the iGEM Team from [https://2012.igem.org/Team:University_College_London University College London]== |
| + | We started collaboration with the iGEM Team [https://2012.igem.org/Team:University_College_London University College London]. They also work with a laccase, the laccase from ''E. coli'' W3110. So we had good requirements to characterize each other’s laccases with the different methods we used to characterize the proteins. Therefore we sent our part <partinfo>BBa_K863005</partinfo> and got the part <partinfo>BBa_K729006</partinfo>. In our experiments we showed that their laccase is expressed and active using our [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Activity_measurements activity test] with ABTS. <br> For more detailed information about the methods we used for characterization and the results have a look on the [https://2012.igem.org/Team:Bielefeld-Germany/Results/london Result Page] or the entry in [http://partsregistry.org/Part:BBa_K729006:Experience Parts Registry]. | ||
| - | |||
| - | |||
| - | |||
| - | + | == Collaboration with the iGEM team from [https://2012.igem.org/Team:SDU-Denmark the Southern University of Denmark]== | |
| - | + | We invited the iGEM Team from the Southern University of Denmark to Bielefeld to share and exchange knowledge and of course to meet another iGEM team. Furthermore they supported us on our event [https://2012.igem.org/Team:Bielefeld-Germany/StreetScience Street Science]. | |
| - | + | You can find all information about our meeting [https://2012.igem.org/Team:Bielefeld-Germany/Collaboration here]. | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | + | ==Collaboration with the iGEM team from the [https://2012.igem.org/Team:LMU-Munich LMU Munich]== | |
| - | + | We sent our plasmids with ECOL <partinfo>BBa_K863005</partinfo> and BHAL <partinfo>BBa_K863020</partinfo> to the [https://2012.igem.org/Team:LMU-Munich LMU Munich iGEM Team]. They asked us in Amsterdam for theese two constructs of us and wanted to express and immobilize them on their ''Bacillus subtilis'' [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins sporobeads]. | |
| - | = | + | <br style="clear: both" /> |
| - | + | ||
| - | + | ==Collaboration with the [https://2012.igem.org/Team:British_Columbia UBC iGEM team]== | |
| - | + | The UBC iGEM team prepared an Intellectual Property Survey, which was answered by our whole team. | |
| - | + | <br> | |
| - | + | <br> | |
| + | ==Collaboration with the TU Munich== | ||
| + | {{Team:TU_Munich/Badge}} | ||
| - | + | ||
| - | + | </div> | |
<html> | <html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| Line 143: | Line 124: | ||
</div> | </div> | ||
| + | |||
| + | |||
| Line 150: | Line 133: | ||
| - | $("#tab ul").tabs("# | + | $("#tab ul").tabs("#judging_site > div", {effect: 'fade', fadeOutSpeed: 400}); |
}); | }); | ||
</script> | </script> | ||
| - | |||
| - | + | ||
| + | </html> | ||
Latest revision as of 02:43, 27 October 2012
Our BioBricks
<groupparts>iGEM012 Bielefeld-Germany</groupparts>
Datapage
How our BioBricks work
Data for our favorite new parts
- <partinfo>K863000</partinfo> - bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and His-tag: This part is used to overexpress the laccase bpul for further purification followed by characterization of enzyme activity and substrate specificity.
- <partinfo>K863005</partinfo> - ecol (laccase from E. coli) with T7 promoter, RBS and His-tag: This enzyme is overexpressed after induction and can be purified by His-tag. Subsequently the laccase can be characterized regarding to enzyme activity and substrate specificity.
- <partinfo>K863204</partinfo> - shuttle vector pECPP11JS for site-directed recombination of genes of interest in yeast: With this part the production and secretion of a protein of interest like laccase is possible.
We have also characterized the following parts
- <partinfo>BBa_K863012</partinfo> - tthl laccase (from T. thermophilus) with constitutive promoter J23100, RBS and His-tag: This part is used for the constitutive expression of tthl for further purification followed by characterization of enzyme activity and substrate specificity.
- <partinfo>BBa_K863022</partinfo> - bhal laccase (from Bacillus halodurans) with constitutive promoter J23100, RBS and His-tag: This part is used for the constitutive expression of bhal for further purification followed by characterization of enzyme activity and substrate specificity.
- <partinfo>BBa_K863207</partinfo> - shuttle vector pECPP11JS for site-directed recombination of TVEL5 laccase in yeast: With this part the production and secretion of TVEL5 is possible.
Collaborations
Collaboration with the iGEM Team from University College London
We started collaboration with the iGEM Team University College London. They also work with a laccase, the laccase from E. coli W3110. So we had good requirements to characterize each other’s laccases with the different methods we used to characterize the proteins. Therefore we sent our part <partinfo>BBa_K863005</partinfo> and got the part <partinfo>BBa_K729006</partinfo>. In our experiments we showed that their laccase is expressed and active using our activity test with ABTS.
For more detailed information about the methods we used for characterization and the results have a look on the Result Page or the entry in [http://partsregistry.org/Part:BBa_K729006:Experience Parts Registry].
Collaboration with the iGEM team from the Southern University of Denmark
We invited the iGEM Team from the Southern University of Denmark to Bielefeld to share and exchange knowledge and of course to meet another iGEM team. Furthermore they supported us on our event Street Science. You can find all information about our meeting here.
Collaboration with the iGEM team from the LMU Munich
We sent our plasmids with ECOL <partinfo>BBa_K863005</partinfo> and BHAL <partinfo>BBa_K863020</partinfo> to the LMU Munich iGEM Team. They asked us in Amsterdam for theese two constructs of us and wanted to express and immobilize them on their Bacillus subtilis sporobeads.
Collaboration with the UBC iGEM team
The UBC iGEM team prepared an Intellectual Property Survey, which was answered by our whole team.
Collaboration with the TU Munich
| 55px | | | | | | | | | | |
 "
"







