Team:Colombia/Parts
From 2012.igem.org
(Prototype team page) |
Sylvita1015 (Talk | contribs) |
||
| (52 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{https://2012.igem.org/User:Tabima}} | |
| - | + | <div class="right_box"> | |
| - | <div | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == '''Parts''' == | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | == '''Toxin- Antitoxin (TA) Modules''' == | ||
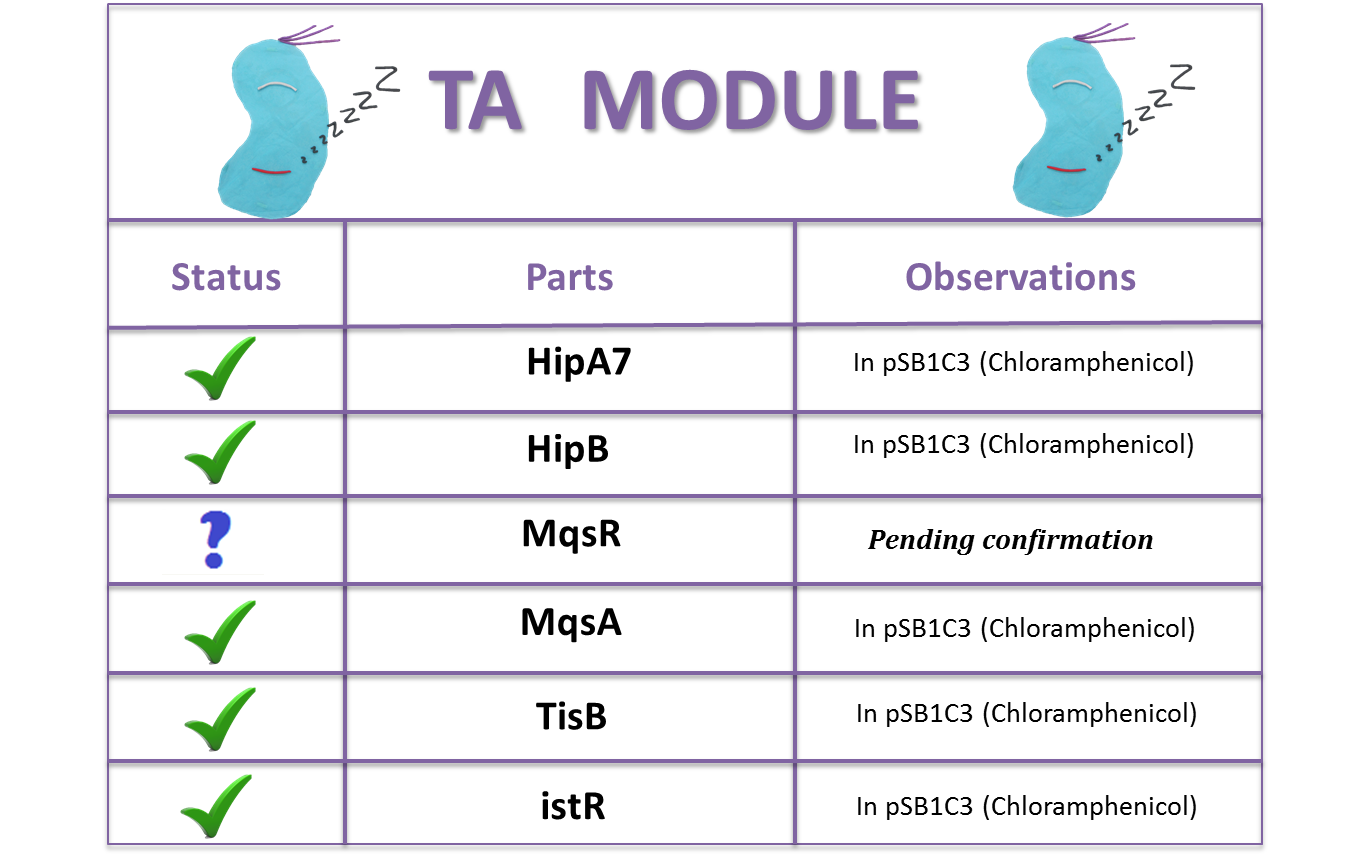
| - | + | == Our Basic Parts == | |
| - | |||
| + | [[File:TA_BioBricks1-2.png|center|550px]] | ||
| - | + | ||
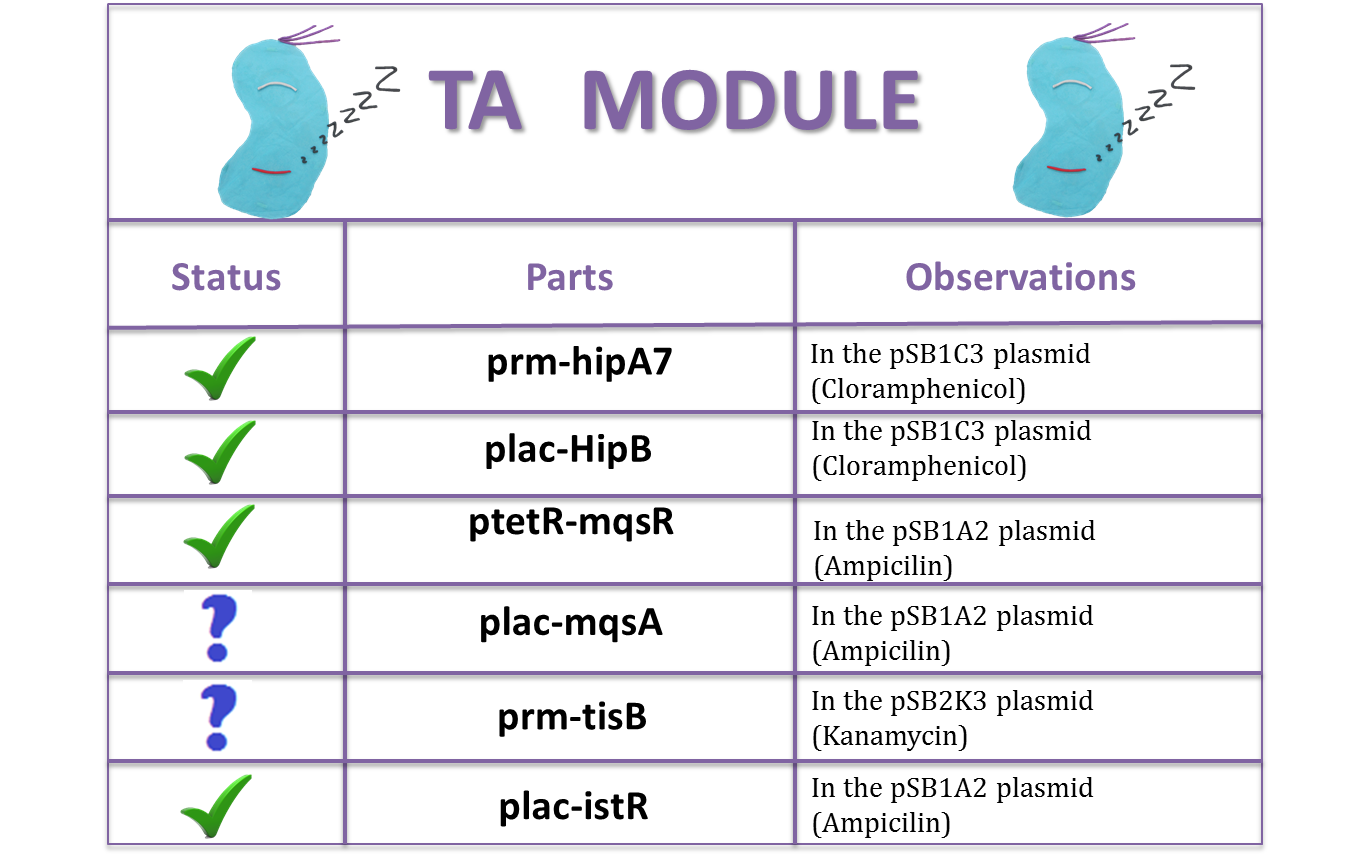
| + | == Our Composite Parts == | ||
| + | |||
| + | [[File:TA_BioBricks3.png|center|550px]] | ||
| + | |||
| + | ='''We send all this parts to the Registry!!!! '''= | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K831000:Design HipA7] | ||
| + | [http://partsregistry.org/Part:BBa_K831003:Design TisB] | ||
| + | [http://partsregistry.org/Part:BBa_K831001:Design HipB] | ||
| + | [http://partsregistry.org/Part:BBa_K831002:Design MqsA] | ||
| + | [http://partsregistry.org/Part:BBa_K831004:Design ''istR'' ] | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K831015 prm-''hipA7''] | ||
| + | [http://partsregistry.org/Part:BBa_K831009 plac-''hipB''] | ||
| + | [http://partsregistry.org/Part:BBa_K831007 ptetR-''mqsR''] | ||
| + | [http://partsregistry.org/Part:BBa_K831014 plac-''istR''] | ||
| + | |||
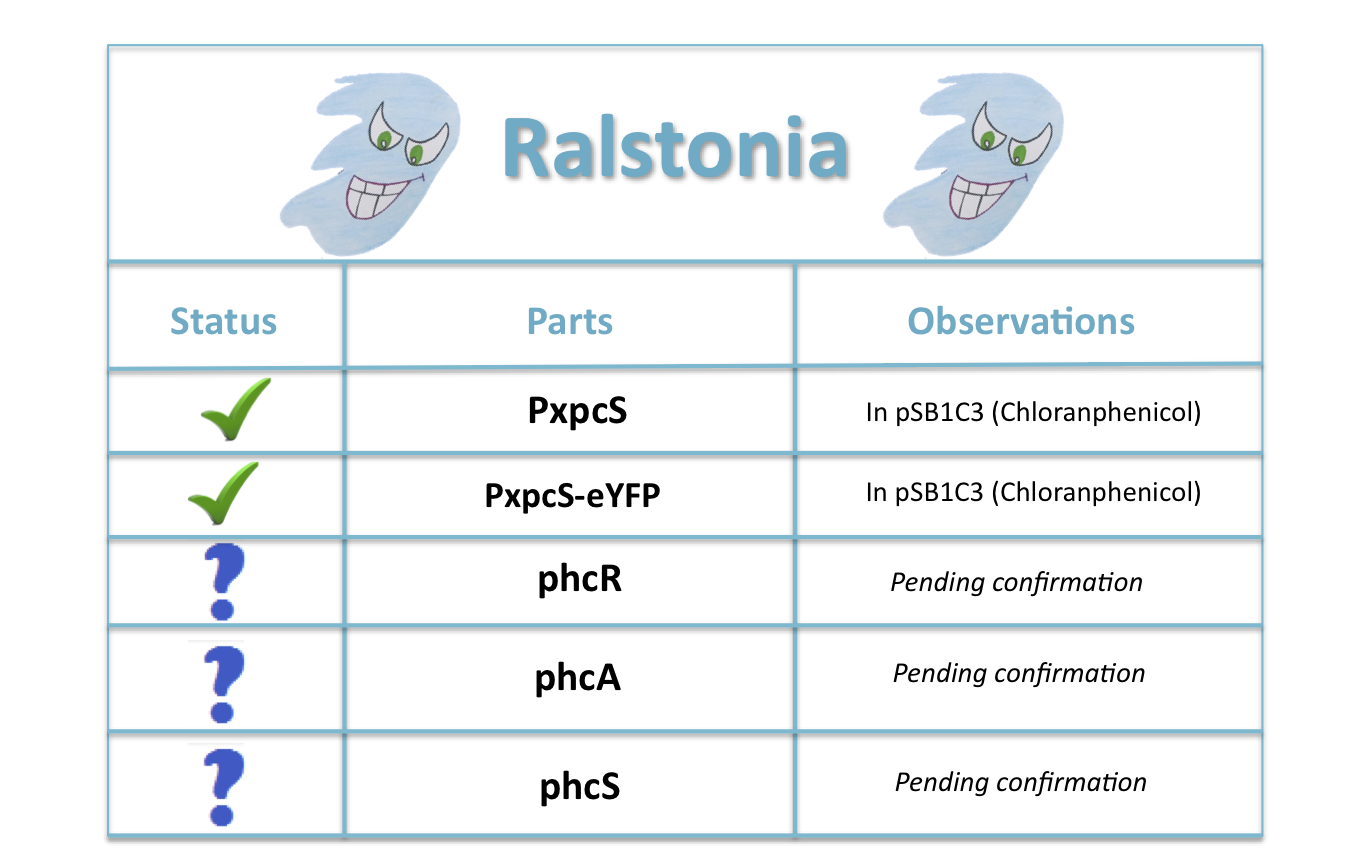
| + | == '''Ralstonia:''' == | ||
| + | |||
| + | [[File:Ralstoniaparts.png|center|550px]] | ||
| + | |||
| + | |||
| + | '''pchS:''' It encodes a histidine kinase sensor of a two-component signal transduction system. Responds to threshold concentrations of the Quorum Sensing unusual signal 3-OH PAME. | ||
| + | |||
| + | '''pchR:''' Receptor of a two-component signal transduction system. Responds to threshold concentrations of the Quorum Sensing unusual signal 3-OH PAME. It is an atypical response regulator that has a histidine kinase as its output domain. Autophosphorilation after 3OH-PAME recognition may have en effect in phcA activation. | ||
| + | |||
| + | '''pchA:'''LysR-type transcriptional regulator that controls expression of many genes and is induced in a density manner, relying on the unusual Quorum Sensing signal 3OH-PAME. | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K831016 '''PxpsR:'''] Promoter region of xpsR (PxpsR) wich integrates different signals including phcA. Expression of genes regulated by XpsR may be correlated with threshold concentrations of 3 OH-PAME. | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K831017 '''PxpsR-eYFP:'''] Fluorescent Reporter to asses the response of PxpsR to different concentrations of 3OH-PAME. | ||
| + | |||
| + | == ''Aliivibrio fischeri'' == | ||
| + | |||
| + | [[File:alis.png|center|500px|]] | ||
| + | |||
| + | The overall model can be seen in figures in [https://2012.igem.org/Team:Colombia/Project/Experiments/Aliivibrio_and_Streptomyces the main page of Aliivibrio/Streptomyces.] | ||
| + | |||
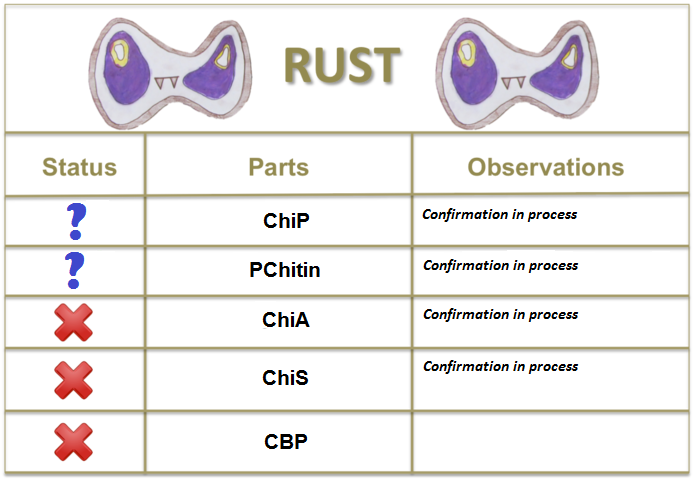
| + | '''chiP:''' [http://www.ncbi.nlm.nih.gov/gene/3279194 Chitoporin] is a specific channel that allows N-acetylglucosamine dimers to cross the outer membrane to the periplasmic space, | ||
| + | |||
| + | '''chiA:''' [http://www.ncbi.nlm.nih.gov/gene/3277358 Chitinase] is an hydrolase that cleaves the glycosidic bonds on the chitin polymer. | ||
| + | |||
| + | '''chiS:''' [http://www.ncbi.nlm.nih.gov/gene/3279287 The histidine-kinase sensor] is part of a two-component signalling system, that activates a promoter, in this case pChitin. | ||
| + | |||
| + | '''cbp:''' [http://www.ncbi.nlm.nih.gov/gene/3279341 Chitin-binding protein] is able to bind the N-acetyl glucosamine dimers in the periplasmic space. The complex CBP+(GlcNAc)2 serves as an activator of the signalling from chiS to the promoter. | ||
| + | |||
| + | '''PChitin:''' This 100bp region upstream of chiP is thought to be activated by the activation of chiS. | ||
| + | |||
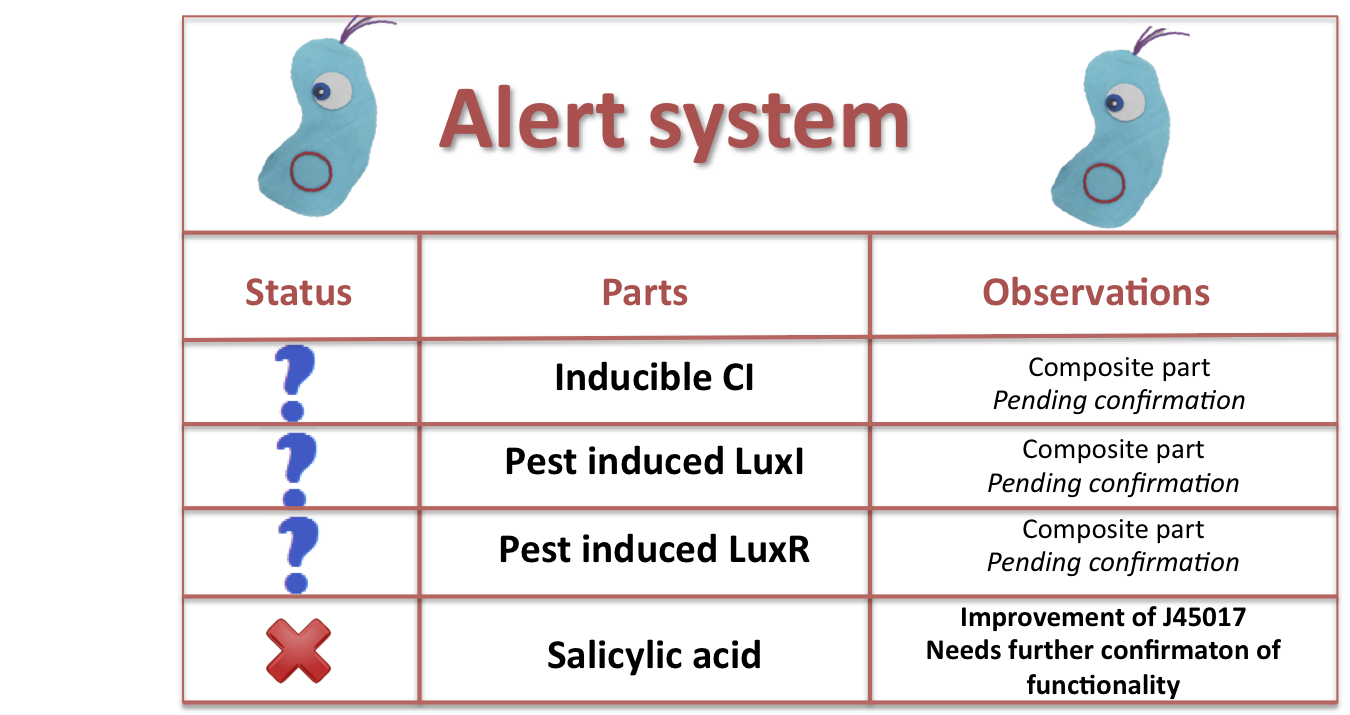
| + | == Alert System: == | ||
| + | |||
| + | [[File:Cutandpasteparts.png|center|600px]] | ||
| + | |||
| + | </div> | ||
Latest revision as of 01:42, 27 October 2012
Template:Https://2012.igem.org/User:Tabima
Contents |
Parts
Toxin- Antitoxin (TA) Modules
Our Basic Parts
Our Composite Parts
We send all this parts to the Registry!!!!
[http://partsregistry.org/Part:BBa_K831000:Design HipA7] [http://partsregistry.org/Part:BBa_K831003:Design TisB] [http://partsregistry.org/Part:BBa_K831001:Design HipB] [http://partsregistry.org/Part:BBa_K831002:Design MqsA] [http://partsregistry.org/Part:BBa_K831004:Design istR ]
[http://partsregistry.org/Part:BBa_K831015 prm-hipA7] [http://partsregistry.org/Part:BBa_K831009 plac-hipB] [http://partsregistry.org/Part:BBa_K831007 ptetR-mqsR] [http://partsregistry.org/Part:BBa_K831014 plac-istR]
Ralstonia:
pchS: It encodes a histidine kinase sensor of a two-component signal transduction system. Responds to threshold concentrations of the Quorum Sensing unusual signal 3-OH PAME.
pchR: Receptor of a two-component signal transduction system. Responds to threshold concentrations of the Quorum Sensing unusual signal 3-OH PAME. It is an atypical response regulator that has a histidine kinase as its output domain. Autophosphorilation after 3OH-PAME recognition may have en effect in phcA activation.
pchA:LysR-type transcriptional regulator that controls expression of many genes and is induced in a density manner, relying on the unusual Quorum Sensing signal 3OH-PAME.
[http://partsregistry.org/Part:BBa_K831016 PxpsR:] Promoter region of xpsR (PxpsR) wich integrates different signals including phcA. Expression of genes regulated by XpsR may be correlated with threshold concentrations of 3 OH-PAME.
[http://partsregistry.org/Part:BBa_K831017 PxpsR-eYFP:] Fluorescent Reporter to asses the response of PxpsR to different concentrations of 3OH-PAME.
Aliivibrio fischeri
The overall model can be seen in figures in the main page of Aliivibrio/Streptomyces.
chiP: [http://www.ncbi.nlm.nih.gov/gene/3279194 Chitoporin] is a specific channel that allows N-acetylglucosamine dimers to cross the outer membrane to the periplasmic space,
chiA: [http://www.ncbi.nlm.nih.gov/gene/3277358 Chitinase] is an hydrolase that cleaves the glycosidic bonds on the chitin polymer.
chiS: [http://www.ncbi.nlm.nih.gov/gene/3279287 The histidine-kinase sensor] is part of a two-component signalling system, that activates a promoter, in this case pChitin.
cbp: [http://www.ncbi.nlm.nih.gov/gene/3279341 Chitin-binding protein] is able to bind the N-acetyl glucosamine dimers in the periplasmic space. The complex CBP+(GlcNAc)2 serves as an activator of the signalling from chiS to the promoter.
PChitin: This 100bp region upstream of chiP is thought to be activated by the activation of chiS.
 "
"