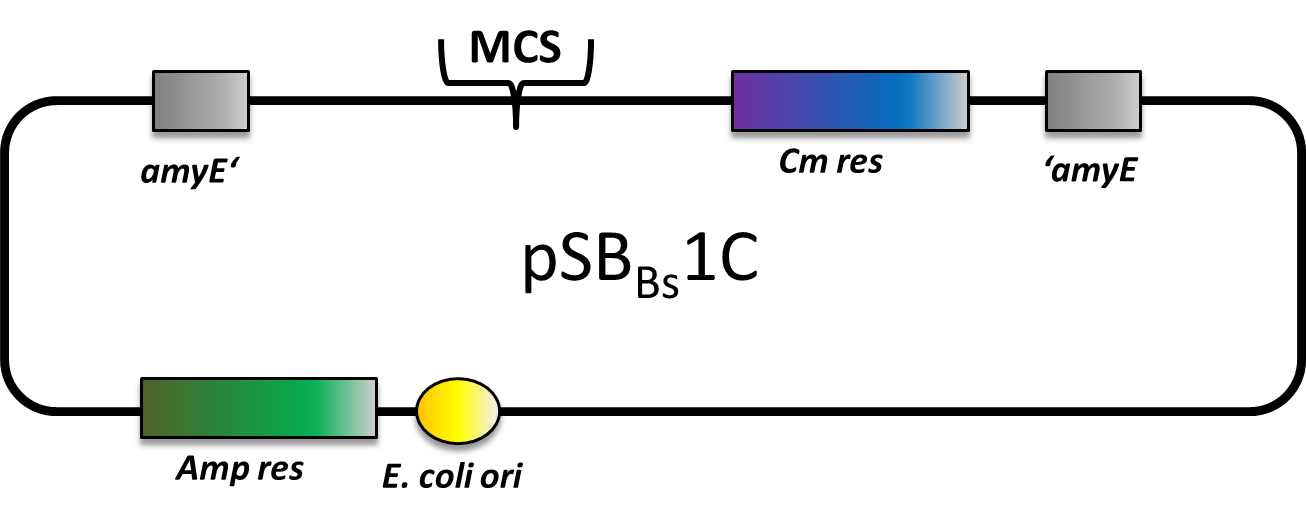|
|
| (285 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | <!-- Include the next line at the beginning of every page --> | | <!-- Include the next line at the beginning of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_culture_tubes.resized.jpg|3}} | | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_culture_tubes.resized.jpg|3}} |
| | + | [[File:Bacillus BioBrick Box banner.resized WORDS.JPG|620px|link=]] |
| | | | |
| - | [[Image:BacillusBioBrickBox.png|150px|right]]
| |
| - | ==''Bacillus'' BioBricks==
| |
| | | | |
| | + | [[Image:BacillusBioBrickBox.png|100px|right|link=]] |
| | | | |
| - | We will create a toolbox of <i>Bacillus</i> BioBricks to contribute to the registry.
| |
| | | | |
| - | This ''<b>B</b>acillus'' <b>B</b>io<b>B</b>rick <b>B</b>ox contains ''Bacillus'' specific:
| |
| | | | |
| - | * '''[[Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]]'''
| + | =='''B<sup>4</sup>''' - 22 core parts for ''Bacillus subtilis''== |
| - | * '''[[Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Promoters|Promoters]]'''
| + | <br> |
| - | * '''[[Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Reporters|Reporters]]'''
| + | <p align="justify">A major goal of our iGEM project is to [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction introduce ''B. subtilis''] as a new chassis for BioBrick-based synthetic biology. For that purpose, we created a toolbox of <i>Bacillus</i> BioBricks to contribute to the registry to make it accessible to many more future iGEM-teams and the entire public microbiology domain! This ''<b>Bacillus'' B</b>io<b>B</b>rick <b>B</b>ox ('''B<sup>4</sup>''') contains the following ''Bacillus'' specific parts:</p> |
| | | | |
| | | | |
| | + | <div class="box"> |
| | + | {| width="100%" |
| | | | |
| - | ==''Bacillus'' Vectors==
| + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Vectors|Vectors]]''' |
| - | | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Promoters|Promoters]]''' |
| - | <p align="justify">Here is a list of all the vectors we cloned and used. Please note that pMAD is not in a BioBrick standard, but it is a useful tool to knock out genes in ''Bacillus subtilis''.</p>
| + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Reporters|Reporters]]''' |
| - | | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Tags|Affinity tags]] |
| - | <p align="justify">For the use of our vectors, please see our [[Team:LMU-Munich/Lab_Notebook/Protocols|Protocols]] page. A general introduction to ''Bacillus subtilis'' and its integrative vectors can be found [[Team:LMU-Munich/Bacillus Introduction|here]]. All vectors have ampicillin as <i>E. coli</i> resistance and RFP as selection marker.</p>
| + | |
| - | | + | |
| - | | + | |
| - | {| class="colored"
| + | |
| - | |- | + | |
| - | !Name
| + | |
| - | !''E. coli'' res.
| + | |
| - | !''B. subt.'' res.
| + | |
| - | !Insertion locus
| + | |
| - | !Description
| + | |
| - | !Derived from
| + | |
| - | !Reference
| + | |
| - | |-
| + | |
| - | !pSB<sub>Bs</sub>1C-<i>lacZ</i>
| + | |
| - | |Amp | + | |
| - | |Cam
| + | |
| - | |amyE
| + | |
| - | |with <i>lacZ</i> reporter gene
| + | |
| - | |pAC6
| + | |
| - | |[http://www.ncbi.nlm.nih.gov/pubmed/11902727 Stülke ''et al''.]
| + | |
| - | |- | + | |
| - | !pSB<sub>Bs</sub>4S
| + | |
| - | |Amp | + | |
| - | |Spec
| + | |
| - | |thrC
| + | |
| - | |empty
| + | |
| - | |pDG1731
| + | |
| - | |[http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury ''et al.'']
| + | |
| - | |-
| + | |
| - | !pSB<sub>Bs</sub>1C
| + | |
| - | |Amp
| + | |
| - | |Cam
| + | |
| - | |amyE
| + | |
| - | |empty
| + | |
| - | |pDG1662
| + | |
| - | |[http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury ''et al.'']
| + | |
| - | |-
| + | |
| - | !pSB<sub>Bs</sub>4S-P<sub><i>Xyl</i></sub>
| + | |
| - | |Amp | + | |
| - | |Spec
| + | |
| - | |thrC
| + | |
| - | |with Xylose-inducible promoter
| + | |
| - | |pXT
| + | |
| - | |[http://www.ncbi.nlm.nih.gov/pubmed/11069659 Derré ''et al.'']
| + | |
| - | |-
| + | |
| - | !pSB<sub>Bs</sub>3C-<i>luxABCDE</i>
| + | |
| - | |Amp
| + | |
| - | |Cam
| + | |
| - | |sacA
| + | |
| - | |with <i>luxABCDE</i> reporter cassette
| + | |
| - | |pAH328
| + | |
| - | |[http://www.ncbi.nlm.nih.gov/pubmed/20709900 Schmalisch ''et al''.]
| + | |
| | |- | | |- |
| - | !pSB<sub>Bs</sub>0K-P<sub><i>spac</i></sub>
| + | |[[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]] |
| - | |Amp | + | |[[File:LMU PromoterIconBC.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Promoters]] |
| - | |Kan | + | |[[File:LMU Reporter.png|60px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Reporters]] |
| - | |replicative | + | |[[File:Proteinaffinitytagbutton.png|50px|link=Team:LMU-Munich/Bacillus_BioBricks#Affinity_Tags]] |
| - | |with IPTG inducible promoter | + | |
| - | |pDG148 | + | |
| - | |http://www.ncbi.nlm.nih.gov/pubmed/11728721 Joseph ''et al.''] | + | |
| | |- | | |- |
| - | !pSB<sub>Bs</sub>2E
| + | |valign="top"|<font size="2"> |
| - | |Amp
| + | pSB<sub>''Bs''</sub>1C<br> |
| - | |MLS
| + | pSB<sub>''Bs''</sub>4S<br> |
| - | |lacA
| + | pSB<sub>''Bs''</sub>2E<br> |
| - | |empty
| + | pSB<sub>''Bs''</sub>1C-''lac''Z<br> |
| - | |pAX01
| + | pSB<sub>''Bs''</sub>3C-''lux''ABCDE<br> |
| - | |[http://www.ncbi.nlm.nih.gov/pubmed/11274134 Härtl ''et al.''] | + | pSB<sub>''Bs''</sub>4S-P<sub>''xyl''</sub><br> |
| | + | pSB<sub>''Bs''</sub>0K-P<sub>''spac''</sub><br> |
| | + | '''Sporo'''vector</font> |
| | + | |valign="top"|<font size="2"> |
| | + | Anderson<br> |
| | + | P<sub>''liaG''</sub><br> |
| | + | P<sub>''veg''</sub><br> |
| | + | P<sub>''lepA''</sub><br> |
| | + | P<sub>''liaI''</sub><br> |
| | + | ''xylR''-P<sub>''xyl''</sub></font> |
| | + | |valign="top"|<font size="2"> |
| | + | ''gfp''<br> |
| | + | ''mkate2''<br> |
| | + | ''lacZ''<br> |
| | + | ''luc+''<br></font> |
| | + | |valign="top"|<font size="2"> |
| | + | Flag<br>His<br>cMyc<br>Strep<br>HA</font> |
| | |- | | |- |
| | |} | | |} |
| | + | </div> |
| | | | |
| | | | |
| - | <p align="justify">The number in the vector's name codes for the insertion locus and the following letter for the <i> Bacillus subtilis </i> resistance gene according to the following table:</p>
| |
| | | | |
| | + | [[File:NEXT.png|right|80px|link=Team:LMU-Munich/Germination_Stop]] [[File:BACK.png|left|80px|link=Team:LMU-Munich/Data/differentiation_tour]] |
| | | | |
| - | {| class="colored"
| |
| - | |-
| |
| - | !Number
| |
| - | !Insertion locus
| |
| - | !Letter
| |
| - | !Resistance
| |
| - | |-
| |
| - | !0
| |
| - | |replicative
| |
| - | !C
| |
| - | |Chloramphenicol
| |
| - | |-
| |
| - | !1
| |
| - | |amyE (amylase)
| |
| - | !E
| |
| - | |MLS (Erythromycin + Lincomycin)
| |
| - | |-
| |
| - | !2
| |
| - | |lacA (laccase ?)
| |
| - | !K
| |
| - | |Kanamycin
| |
| - | |-
| |
| - | !3
| |
| - | |sacA (saccase?)
| |
| - | !S
| |
| - | |Spectinomycin
| |
| - | |-
| |
| - | !4
| |
| - | |thrC (threonine C)
| |
| - | |
| |
| - | |
| |
| - | |-
| |
| | | | |
| - | |-
| |
| - | |}
| |
| | | | |
| | | | |
| - | The concentrations of the antibiotics and the insertion tests can be found in our [https://2012.igem.org/Team:LMU-Munich/Lab_Notebook/Protocols Protocol] section.
| |
| | | | |
| - | The promoters we submit are:
| |
| | | | |
| - | Pveg, PliaI, PliaG, plepA, Pxyl, Pxyl+XylR
| |
| | | | |
| - | <p align="justify">We also tested the [http://partsregistry.org/Promoters/Catalog/Anderson Anderson promoter collection] in ''Bacillus subtilis'' with pSB<sub>Bs</sub>3C-<i>luxABCDE</i>. The data will be online in our [https://2012.igem.org/Team:LMU-Munich/Data Data] section soon.</p>
| |
| | | | |
| - | <p align="justify">Furthermore we plan to submit reporter genes optimized for ''Bacillus subtilis'', namely GFP, lacZ, luc and mKate. </p>
| |
| | | | |
| - | Useful for the registry are also 5 tags in Freiburg Standard (cMyc, 10His, Flag, Strep, HA).
| + | <p align="justify">Since ''Bacillus subtilis'' is not an organism commonly used in iGEM, please check out our [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction Introduction] to learn more about it.</p> |
| | | | |
| - | More details to come soon.
| + | <div class="box"> |
| | + | ==''Bacillus'' Vectors [[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]]== |
| | + | {| "width=100%" style="text-align:center;" style="align:right"| |
| | + | |<p align="justify">We have generated a suite of BioBrick-compatible vectors: three empty insertional backbones with different antibiotic resistances and integration loci, two reporter and two expression vectors.</p> |
| | + | |[[File:LMU-Munich-PSBBs1C.png|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Vectors]] |
| | + | |- |
| | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Vectors]] |
| | + | |} |
| | + | </div> |
| | | | |
| | + | <div class="box"> |
| | + | ==''Bacillus'' Promoters [[File:LMU PromoterIconBC.png|100px]]== |
| | + | {| "width=100%" style="align:right"| |
| | + | | |
| | + | <p align="justify">To provide a set of promoters of different strength we characterized several promoters in ''Bacillus subtilis''. Both constitutive and inducible promoters are covered.</p> |
| | + | | |
| | + | [[File:Promoters overview.png|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Promoters]] |
| | + | |- |
| | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Promoters]] |
| | + | |} |
| | + | </div> |
| | | | |
| - | ==''Bacillus'' Promoters== | + | <div class="box"> |
| | + | ==''Bacillus'' Reporters [[File:LMU Reporter.png|50px]]== |
| | + | {| "width=100%" style="align:right"| |
| | + | | |
| | + | <p align="justify">We designed and codon-optimized a set of reporters that are commonly used in ''B. subtilis''.</p> |
| | + | | |
| | + | [[File:LMU GFP.jpg|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Reporters]] |
| | + | |- |
| | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Reporters]] |
| | + | |} |
| | + | </div> |
| | | | |
| - | <p align="justify">Another goal is to produce promoters of ''Bacillus subtilis'' in BioBrick mode and to evaluate them. These well-defined promoters will then be part of the [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks ''Bacillus'' BioBrickBox] which we will send to the registry, but they can also be useful in our project [https://2012.igem.org/Team:LMU-Munich/Project '''Bead'''zillus] to express our [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins fusion crust proteins] on the outside of our spores. Therefore we will use different promoters which are the constitutive promoters from the [http://partsregistry.org/Promoters/Catalog/Anderson Anderson collection] from the Parts Registry, the constitutive promoters P<sub>''liaG''</sub>, P<sub>''veg''</sub> and P<sub>''lepA''</sub> from ''B. subtilis'' as well as the inducible promoter P<sub>''liaI''</sub> from ''B. subtilis''. For the characterization of the different promoters we will clone them upstream of reporter genes (lux operon, lacZ) to measure their activity in ''B. subtilis''. Therefore we use e.g. the two reporter vectors from ''B. subtilis'' which are also part of the [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks ''Bacillus'' BioBrickBox]. One of the reporter vectors pSB<sub>Bs</sub>3C-<i>luxABCDE</i> contains the ''lux'' operon as a reporter. This is why the activity of the promoter can be measured as luminescence with the plate reader (BioTek) which is a result of the expression of the luciferase. The second reporter vector used for the evaluation of the promoters is pSB<sub>Bs</sub>1C-''lacZ'' which contains the reporter gene ''lacZ''. The promoter activity leads to the production of the enzyme beta-galactosidase which cleaves the substrate ONPG. The product is detectable with the photometer and refers to the promoter activity.</p> | + | <div class="box"> |
| - | | + | ==Affinity Tags [[File:Proteinaffinitytagbutton.png|50px]]== |
| - | | + | {| "width=100%" style="align:right"| |
| - | === Promoter Evaluation === | + | | |
| - | | + | <p align="justify">We synthesized 5 affinity tags for protein purification. They all are designed in Freiburg standard with an optimized ribosome binding site upstream. We have not yet tested our tags.</p> |
| - | <p align="justify">To get a set of promoters with different strength we chose several promoters. They can be divided in three different groups: the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the Parts Registry, the constitutive promoters P''liaG'', P''veg'' and P''lepA'' from ''B. subtilis'', and the inducible promoter P''liaI'' from ''B. subtilis''. For the characterization of the different promoters, they were all cloned upstream of reporter genes (lux operon, lacZ) to measure their activity in ''B. subtilis'' by regarding the gene expression of the reporter gene.</p>
| + | |
| - | | + | |
| - | | + | |
| - | ====Anderson promoters====
| + | |
| - | | + | |
| - | <p align="justify">The first group of promoters evaluated are the promoters of the Anderson collection which we call ''Anderson promoters''. They have already been measured in ''Escherichia coli'' and they all showed a constitutive behavior but with a different strength. In this project, these Anderson promoters were measured in ''B. subtilis''. As they have already been evaluated in ''E. coli'', they were already in the Parts Registry in the vector where they are fused to the ''Red fluorescent protein'' (RFP). For evaluation they were cut out of the vector and then first cloned in BioBrick standard in the empty vector pSB1C3 to send them to the registry. In addition, eleven (J23100,J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) of the nineteen promoters of the Anderson collection were successfully cloned in the reporter vector pSB<sub>Bs</sub>3C-<i>luxABCDE</i> from the BioBrickBox containing the ''lux'' operon as a reporter for promoter activity. The gene expression which correlates to the promoter activity leads to the expression of the ''lux'' operon with the luciferase. The luminescence which is produced by the luciferase can be measured with the plate reader (BioTek). To measure the activity not only with the lux reporter operon, four promoters were cloned into the reporter vector pSB<sub>Bs</sub>1C-''lacZ'' to do beta-galactosidase assays and then to compare the results of the strength of these promoters in ''B. subtilis''.</p>
| + | |
| - | | + | |
| - | | + | |
| - | ====Constitutive promoters from ''B. subtilis''====
| + | |
| - | | + | |
| - | <p align="justify">The second group of promoters are constitutive promoters from ''B. subtilis''. We will evaluate the promoters P<sub>''liaG''</sub>, P<sub>''veg''</sub> and P<sub>''lepA''</sub>. Therefore we will use the reporter vectors pSB<sub>Bs</sub>3C-<i>luxABCDE</i> and pSB<sub>Bs</sub>1C-''lacZ'' to evaluate the promoters with different reporters, the ''lux'' operon and the ''lacZ'' gene. Therefore the promoters will be amplified from the genome of ''B. subtilis'' with primers that conatin the restriction sites of the BioBrick standard. Then these constitutive promoters will be cloned into the empty vector pSB1C3 to send them to the registry as well as the two reporter vectors to evaluate their strength in ''B. subtilis''.</p>
| + | |
| - | | + | |
| - | | + | |
| - | ====Inducible promoters from ''B. subtilis''====
| + | |
| - | | + | |
| - | <p align="justify">The last group of promoters consists of inducible promoters of ''B. subtilis'' e.g. P''<sub>liaI</sub>''. They are useful to decide when to turn on gene expression because these promoters need an inducer to start transcription. P''<sub>liaI</sub>'' can be induced by antibiotics which interact with the lipidII cycle, e.g. bacitracin. In this project this promoter is evaluated like the constitutive promoters with the reporter vector pSB<sub>Bs</sub>3C-<i>luxABCDE</i> which contains the ''lux'' operon as a reporter and with the vector pSB<sub>Bs</sub>1C-''lacZ'' which contains the ''lacZ'' reporter gene. Therefore the promoters will be amplified from the genome of ''B. subtilis'' with primers that contain the restriction sites of the BioBrick standard. Then these inducible promoters will be cloned into the empty vector pSB1C3 to send them to the registry as well as the two reporter vectors to evaluate their strength in ''B. subtilis''. To turn the promoter on we have to add an inducer.</p>
| + | |
| - | | + | |
| - | | + | |
| - | ==''Bacillus'' Reporters==
| + | |
| - | | + | |
| - | <p align="justify">We designed some reporters that are commonly used in ''B. subtilis'' or are codon optimized versions of popular reporter genes. All reporters have a modified iGEM Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RCF 25]) pre- and suffix for assembly of in-frame fusion proteins. Our prefix also includes the ''B. subitlis'' optimized RBS.</p>
| + | |
| - | | + | |
| - | prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
| + | |
| - | | + | |
| - | suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
| + | |
| - | | + | |
| - | | + | |
| - | *'''GFP'''
| + | |
| - | <p align="justify">We took a ''gfp'' derivate of the ''Bacillus subtilis'' plasmids pGFPamy and added the BioBrick compatiple pre- and suffix of the Freiburg standard (Assembly 25).</p>
| + | |
| - | | + | |
| - | | + | |
| - | *'''mKate'''
| + | |
| - | <p align="justify">We synthesized this ''rfp'' derivate with a codon-optimized version for the use in ''B. subtilis'' with pre- and suffix of the Freiburg standard</p> | + | |
| - | | + | |
| - | | + | |
| - | *'''LacZ'''
| + | |
| - | [[File:LacZ plate.png|''lacZ'' under the control of Pspac in pSBC3. Plate induced with Xylose (ng/µl)|thumb|300px|left]]
| + | |
| - | | + | |
| - | <p align="justify">This lacZ gene is derived from the ''Bacillus'' reporter vector pAC6. It is constructed in the Freiburg Standard (Assembly 25) for in-frame fusion proteins. It also includes a Shine-Dalgarno Sequence optimized for ''Bacillus subtilis'' translation.</p>
| + | |
| - | | + | |
| - | BioBrick Name: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 BBa_K823019]
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | *'''luc'''
| + | |
| - | | + | |
| - | <p align="justify">We synthesized this luciferase for luminescence assays with luciferin as substrate.</p>
| + | |
| - | [[File:Luciferin reaction mod.png|thumb|center|400px]]
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | '''Project Navigation'''
| + | |
| - | | + | |
| - | {|
| + | |
| - | |<font color="#FFFFFF">+--+--+--+--+--+--+--</font>
| + | |
| - | |<font color="#FFFFFF">+--+--+--+--+--</font>
| + | |
| - | |<font color="#FFFFFF">+--+--+--+--+--+--</font>
| + | |
| - | |<font color="#FFFFFF">+--+--+--+--+--+--</font>
| + | |
| | |- | | |- |
| - | |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Tags]] |
| - | <html>
| + | |} |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction">
| + | </div> |
| - | <img src="https://static.igem.org/mediawiki/2012/8/8d/Bacilluss_Intro.png" height=70"/> | + | <br> |
| - | </a>
| + | <br> |
| - | <font size="1">
| + | <br> |
| - | </font>
| + | <br> |
| - | <br /> | + | <div class="box"> |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction"><font size="1" face="verdana">Bacillus<BR>Intro</font></a>
| + | ====Project Navigation==== |
| - | </html>
| + | {| width="100%" align="center" style="text-align:center;" |
| - | | + | |[[File:Bacilluss_Intro.png|100px|link=Team:LMU-Munich/Bacillus_Introduction]] |
| - | |<html>
| + | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks">
| + | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] |
| - | <img src="https://static.igem.org/mediawiki/2012/7/78/BacillusBioBrickBox.png" height=70"/>
| + | |[[File:GerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] |
| - | </a>
| + | |- |
| - | <font size="1">
| + | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] |
| - | </font>
| + | |[[Team:LMU-Munich/Bacillus_BioBricks|<font size="2" face="verdana">'''''Bacillus'''''<BR>'''B'''io'''B'''rick'''B'''ox</font>]] |
| - | <br />
| + | |[[Team:LMU-Munich/Spore_Coat_Proteins|<font size="2" face="verdana">'''Sporo'''beads</font>]] |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks"><font size="1" face="verdana">Bacillus<BR>BioBrickBOX</font></a>
| + | |[[Team:LMU-Munich/Germination_Stop|<font size="2" face="verdana">'''Germination'''<BR>STOP</font>]] |
| - | </html>
| + | |
| - | | + | |
| - | |<html> | + | |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins">
| + | |
| - | <img src="https://static.igem.org/mediawiki/2012/c/c1/SporeCoat.png" height=70"/>
| + | |
| - | </a>
| + | |
| - | <font size="1">
| + | |
| - | </font>
| + | |
| - | <br /> | + | |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins"><font size="1" face="verdana">SporeCoat<BR>FusionProteins</font></a>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | |<html> | + | |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Germination_Stop">
| + | |
| - | <img src="https://static.igem.org/mediawiki/2012/f/f6/GerminationSTOP.png" height=70"/> | + | |
| - | </a>
| + | |
| - | <font size="1">
| + | |
| - | </font>
| + | |
| - | <br />
| + | |
| - | <a href="https://2012.igem.org/Team:LMU-Munich/Germination_Stop"><font size="1" face="verdana">Germination<BR>STOP</font></a>
| + | |
| - | </html>
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| | | | |
| | <!-- Include the next line at the end of every page --> | | <!-- Include the next line at the end of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Footer}} | | {{:Team:LMU-Munich/Templates/Page Footer}} |














 "
"