Team:Peking/Project/Luminesensor/Characterization
From 2012.igem.org
Spring zhq (Talk | contribs) |
|||
| (79 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
</script> | </script> | ||
<div class="PKU_context floatR first"> | <div class="PKU_context floatR first"> | ||
| - | <h3 | + | <h3 id="title1">Sensitivity</h3> |
<p> | <p> | ||
<b>1. Set-up & Brief Procedure</b><br/> | <b>1. Set-up & Brief Procedure</b><br/> | ||
| - | We tested the sensitivity of <i>Luminesensor</i> by | + | |
| + | |||
| + | |||
| + | We tested the sensitivity of the <i>Luminesensor</i> by examining the light-dependent transcriptional activity of a GFP-ssrA reporter. ssrA is a protein tag that induces fast degradation of protein, which in our case facilitated the observation of transcriptional activity. Based on the consideration of guaranteed accuracy and precision, our setup (Figure 1) is shown below:</p> | ||
| + | |||
| + | |||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="" alt="Figure 2" /> | + | <img src="/wiki/images/a/a4/Peking2012_luminesensor_sen_div.jpg" alt="Figure 2" style="width:500px"/> |
| - | <p class="description">Figure | + | <p class="description">Figure 1. The setup we utilized to characterize the sensitivity of cells harboring the <i>Luminesensor</i></p> |
</div> | </div> | ||
| - | <p> | + | |
| + | |||
| + | <p>Our setup consisted of three central parts: light source, incubator, and 48-well plate. On account of high sensitivity, it is necessary to protect the system from the preventable light exposure with the purpose of acquiring the accurate results which is the true reflection of our sensitivity. In order to solve the problems, we focus on two foremost aspects: 1) utilizing attenuators to weaken the light intensity, and 2) using tin foil to avoid light leakage. | ||
| + | |||
| + | In our experiments, illumination with different light intensity conditions at 460nm peak light from blue LED arrays for 16 hours show marked light-depressed reporter gene transcription, which indicates that under different blue light exposure conditions, there was hardly any light-induced reporter gene transcriptional activity. But when in the dark environment (packaged with three layers of aluminum foil), our systems showed extremely high GFP expression (Figure 2).</p> | ||
| + | |||
| + | |||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src=" | + | <img src="/wiki/images/f/fa/Peking2012_gfp_expression_2.jpg" alt="Figure 3" /> |
| - | <p class="description">Figure | + | <p class="description">Figure 2. GFP expression was repressed by Blue LED light and presented normally in the dark.</p> |
</div> | </div> | ||
| - | |||
| - | |||
| - | Cells exposed to different light intensity expressing <i>Luminesensor</i> showed manifest light-repressed reporter gene transcription. As shown in | + | <p>There are a few plausible and conceivable reasons for why our system possesses such high sensitivity that can sense unbelievably weak light intensity. Firstly, it includes the unusually stable, photo-activated state of VVD, which causes the system to be extremely sensitive to light. Secondly, without relying on the addition of chromophores, which might impede the growth of cells, our system can directly regulate gene expression without trade-off.<br/><br/> |
| + | |||
| + | |||
| + | |||
| + | <b>2. Quantitative Result</b><br/> | ||
| + | |||
| + | |||
| + | Cells exposed to different light intensity expressing the <i>Luminesensor</i> showed manifest light-repressed reporter gene transcription. As shown in Figure 3, all of the cells with dissimilar attenuators showed incredible repression efficiency. <p> | ||
| + | |||
| + | |||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="/wiki/images/a/a8/Peking2012_Luminesensor_sensitivity_2.png" alt="Figure 4" /> | + | <img src="/wiki/images/a/a8/Peking2012_Luminesensor_sensitivity_2.png" alt="Figure 4" style="width:500px"/> |
| - | <p class="description">Figure | + | <p class="description">Figure 3. luminance attenuation using different attenuators could also be sensed by the <i>Luminesensor</i>, which is much dimmer than natural light.</p> |
</div> | </div> | ||
| - | <p>It proves that once the cells are exposed to natural light, the transcription of reporter | + | |
| + | <p>It proves that once the cells are exposed to natural light, the transcription of reporter genes will be strongly repressed, although still present as a dose response. Besides, in the negative control group, which was entirely in the dark state, the expression of GFP ran up to a high degree of 50,000. As a matter of fact, as you can see later in "<a href="https://2012.igem.org/Team:Peking/Project/Communication/Results">Results of light communication between cells</a>", when we serially diluted light-emitting cells which express bacterial luciferase, the cells expressing the <i>Luminesensor</i> present significant dose response. Taking everything into account, our <i>Luminesensor</i> does possess high sensitivity across several orders of magnitude. <br/><br/> | ||
| + | </p> | ||
</div> | </div> | ||
<div class="PKU_context floatR"> | <div class="PKU_context floatR"> | ||
| - | <h3 | + | <h3 id="title2">Orthogonality Test</h3> |
<p> | <p> | ||
| - | |||
| - | Two sections of testing expriments were carried out | + | Two sections of testing expriments were carried out. GFP was selected as a reporter and was fused downstream to two promoters (psulA408 and precA408) controlled by the <i>Luminesensor</i> with the 408 mutation and to two promoters (psulA and precA) controlled by the <i>Luminesensor</i> without the mutation. GFP expression is expected to have a negative relation with repression activity. To be more specific, higher level of green fluorescent indicates weaker repression effect; lower expression of GFP stands for stronger repression. <br/><br/> |
| - | <b>Experimental Design</b><br/>To prove the orthogonality, facts that LexA408-VVD and endogenous LexA work totally independently are needed. Considering practical efficiency, | + | |
| - | <ul><li>1. | + | |
| - | </li><li>2. LexA408-VVD efficiently represses its target | + | |
| + | <b>1. Experimental Design(Figure 4)</b><br/> | ||
| + | |||
| + | To prove the orthogonality, data and facts to prove that LexA408-VVD and the endogenous LexA work totally independently are needed. Considering the practical efficiency, two points of evidence were to be collected:</p> | ||
| + | <ul><li>1. Promoters psulA and precA are repressed in wild-type <i>Ecoli</i> strains while promoters psulA408 and precA408 are not blocked in wild-type strains; | ||
| + | |||
| + | </li><li>2. LexA408-VVD-<i>Luminesensor</i> efficiently represses its target under blue illumination, while it does not repress targets in total darkness. | ||
</li></ul> | </li></ul> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="" alt="Figure 5" /> | + | <img src="/wiki/images/3/39/Peking2012_demonstration_of_orthogonal_result.png" alt="Figure 5" style="width:480px;"/> |
| - | <p class="description">Figure | + | <p class="description">Figure 4. Shows observable color of plates where cell cultures growing under totally dark or light conditions. Different colors are used to indicate three levels of GFP expression.</p> |
</div> | </div> | ||
| - | <p><b>Detailed Methods</b><br/>Experiments were carried out in parallel | + | <p><b>2. Detailed Methods</b><br/> |
| + | |||
| + | |||
| + | |||
| + | Experiments were carried out in parallel -- taking photos of the plates as visual evidence and measuring GFP to quantify the effect. Detailed protocols are as follows:<p> | ||
| + | |||
<ul><li>1. Visible display: | <ul><li>1. Visible display: | ||
</li><li> | </li><li> | ||
| - | <ul><li>Inoculate a single colony into 1.5ml centifuge tube, shaking cultivate cells at 250 rpm/min for 2-3 h at 30℃, then the cell culture | + | <ul><li>Inoculate a single colony into 1.5ml centifuge tube, shaking cultivate cells at 250 rpm/min for 2-3 h at 30℃, then the cell culture is ready for streaking; |
| - | </li><li>Streak cell culture onto | + | </li><li>Streak cell culture onto respective plates; |
| - | </li><li> | + | </li><li>Incubate in total darkness and under light state at 30℃ for 1 or 2 days; |
| - | </li><li>Take photos of | + | </li><li>Take photos of correct plates; |
</li></ul> | </li></ul> | ||
</li><li>2. Data collection: | </li><li>2. Data collection: | ||
</li><li> | </li><li> | ||
| - | <ul><li>Inoculate a proper colony | + | <ul><li>Inoculate a proper single colony into liquid media; |
| - | </li><li> | + | </li><li>Cultivate cells in a shaking state until platform stage at 30 ℃; |
| - | </li><li>Dilute | + | </li><li>Dilute cell cultures with selective media at a ratio of 1:500; |
| - | </li><li> | + | </li><li>Cultivate cells in a shaking state until the stationary stage at 30 ℃ in complete darkness or under blue light; |
| - | </li><li>Harvest cells by centrifugation, | + | </li><li>Harvest cells by centrifugation, then re-suspend pellets in PBS; |
| - | </li><li> | + | </li><li>Measure GFP expression with 96-well-microplate reader; |
</li></ul> | </li></ul> | ||
</li> | </li> | ||
</ul> | </ul> | ||
| - | <p><b>Results</b><br/>Below is a collage of our plates. The plates are placed in groups at 30℃ in either total | + | <p><b>3. Results</b><br/> |
| + | |||
| + | Below is a collage of our plates. The plates are placed in groups at 30℃ in either total darkness or under blue-light illumination. Visual results fit well with Figure 4. Visual appearance of the plates were positive evidence for us.</p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="" alt="Figure 6" /> | + | <img src="/wiki/images/4/49/Peking2012_orthogonality_plate_group1_correction.png" alt="Figure 6(1)." style="width:500px;"/> |
| - | <p class="description">Figure | + | <p class="description">Figure 5(1). Plate streaked with wild type <i>E.coli</i> transformed with wild type <i>Luminesensor</i> and GFP fused to LexA responsive promoters, and cultivated in the dark (A), or under light (B). Evidently there is no GFP expression, indicating that endogenous LexA proteins are capable of fully repress the expression of our wild type reporter gene, rendering our <i>Luminesensor</i> system no longer light-controllable. </p> |
</div> | </div> | ||
| - | |||
<div class="floatC"> | <div class="floatC"> | ||
| - | + | <img src="/wiki/images/8/88/Peking2012_orthogonality_plate_group2.png" alt="Figure 6(2)." style="width:500px;"/> | |
| - | + | <p class="description">Figure 5(2). Plate streaked with wild type <i>E.coli</i> transformed with GFP gene fused to the downstream of psulA408 (A) and precA408 (B). Evidently GFP is perfectly expressed, indicating endogenous LexA proteins are unable to bind to psulA408 and precA408, which suggested that the 408 promoter system is orthogonal to the endogenous SOS system.</p> | |
</div> | </div> | ||
| - | + | <div class="floatC"> | |
| - | + | <img src="/wiki/images/e/e7/Peking2012_orthogonality_plate_group3.png" alt="Figure 6(3)." style="width:500px;"/> | |
| - | + | <p class="description">Figure 5(3). Plate streaked with wild type <i>E.coli</i> transformed with mutated luminesensor and precA408-GFP or psulA408-GFP reporter gene and cultivated in the dark or under light.(A: luminesensor + precA408-GFP, cultivated in the dark. B: luminesensor + precA408-GFP, cultivated under light. C: luminesensor + psulA408-GFP, cultivated in the dark. D: luminesensor + psulA408-GFP, cultivated under light.) Just as we predicted, the reporter gene is expressed in the dark and supressed by our luminesensor under light. This clearly proved that our 408 system is orthogonal to bacteria SOS system while retaining light-controllability.</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | <p> | + | |
| + | <p>Apart from those photos, more accurate results were obtained with Fluorescent Microplate Reader. Below are histograms based on our experimmental data. They clarify how fluoresence changes with lighting condition in different strains(Figure 6).</p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="" alt="Figure | + | <img src="/wiki/images/4/44/Peking2012_precA408-GFP_with_axis_correction1.png" alt="Figure 7" style="width:600px;" /> |
| - | <p class="description">Figure | + | <p class="description">Figure 6(1). GFP expression level under control of precA408 promoter. Column 1 shows expression of precA-GFP reporter gene when transformed into wild-type <i>E.coli</i> without luminesensor. The high expression level indicates that the 408 reporter system is not interfered by endogenous LexA protein. The column 2 and column 3 are expression level of precA-GFP reporter gene co-transformed with our 408 form luminesensor into wild-type <i>E.coli</i>, and cultivated under light (column 2), or under light (column 3). The low expression level under light and hign expression level in the dark indicate that our 408 luminesensor system is totally light-controllable, and possessing a high dynamic range.</p> |
</div> | </div> | ||
| + | <div class="floatC"> | ||
| + | <img src="/wiki/images/9/95/Peking2012_psulA408-GFP_fluorescence_with_axis1.png" alt="Figure 7" style="width:600px;" /> | ||
| + | <p class="description">Figure 6(2). GFP expression level under control of psulA408 promoter. Every column shows data acquired under conditions same as its conterpart in Figure 7(1). The only difference is that the precA408 promoter has been replaced by psulA408 promoter.</p> | ||
| + | </div> | ||
| + | <p>It repeatedly turned out that: | ||
| + | <br /><br /> | ||
| + | 1. reporters under wild-type promoters (psulA and precA) hardly expressed in either illuminated or dark state; their colonies are yellow. In contrast, GFPs fused to promoters psulA408 and precA408 were not blocked in strains, thus they looks bright green. | ||
| + | <br /><br /> | ||
| + | 2. BL21 (DE3) strains expressing both the <i>Luminesensor</i> and its reporters shifted swiftly and sensitively from one states to the other, giving high definition output including lighting condition. | ||
| + | <br /><br /> | ||
| + | To sum up, LexA408-VVD-<i>Luminesensor</i> works independently of host genetic context. | ||
| + | </p> | ||
</div> | </div> | ||
| + | |||
<div class="PKU_context floatR"> | <div class="PKU_context floatR"> | ||
| - | + | <h3 id="title5">Reference</h3> | |
| - | + | <p></p> | |
| - | </ | + | <ul class="refer"><li id="ref1"> |
| - | < | + | 1. Wang, X., Chen, X. & Yang, Y.(2012). spatiotemporal control of gene expression
by a light-switchable transgene system. <i>Nat. Methods</i>, 9: 266: 269 |
| - | + | </li><li id = "ref2"> | |
| - | < | + | 2. Farrell, C.M., Grossman, A.D., and Sauer., R.T.(2005). Cytoplasmic degradation of ssrA-tagged proteins. <i>Mol. Microbiol.</i>, 57: 1750: 61 |
| + | </li><li id = "ref3"> | ||
| + | 3. Roche, E.D., and Sauer., R.T.(2001). Identification of Endogenous SsrA-tagged Proteins Reveals Tagging at Positions Corresponding to Stop Codons. <i>J. Biol. Chem.</i>, 276: 28509: 28515 | ||
| + | </li><li id = "ref3"> | ||
| + | 4. Dimitrova,D., <i>et al.</i>(1997). A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in <i>Escherichia coli</i>.<i>Mol. Gen. Genet.</i>, 257: 205: 212 | ||
| + | </li><li id = "ref3"> | ||
| + | 5. Voigt., C.A.(2006). Genetic parts to program bacteria. <i>Curr. Opin. Biotechnol.</i>, 17: 548: 557 | ||
| + | </li><li id = "ref3"> | ||
| + | 6. Thliveris, A.T., Mount., D.W.(1992). Genetic identification of the DNA binding domain of <i>Escherichia coli</i> LexA protein. <i>Proc. Natl. Acad. Sci. USA</i>, 89 (10): 4500: 4504 | ||
| + | </li></ul> | ||
</div> | </div> | ||
| - | + | </html>{{Template:Peking2012_Color_Epilogue}} | |
Latest revision as of 13:20, 26 October 2012
Sensitivity
1. Set-up & Brief Procedure
We tested the sensitivity of the Luminesensor by examining the light-dependent transcriptional activity of a GFP-ssrA reporter. ssrA is a protein tag that induces fast degradation of protein, which in our case facilitated the observation of transcriptional activity. Based on the consideration of guaranteed accuracy and precision, our setup (Figure 1) is shown below:

Figure 1. The setup we utilized to characterize the sensitivity of cells harboring the Luminesensor
Our setup consisted of three central parts: light source, incubator, and 48-well plate. On account of high sensitivity, it is necessary to protect the system from the preventable light exposure with the purpose of acquiring the accurate results which is the true reflection of our sensitivity. In order to solve the problems, we focus on two foremost aspects: 1) utilizing attenuators to weaken the light intensity, and 2) using tin foil to avoid light leakage. In our experiments, illumination with different light intensity conditions at 460nm peak light from blue LED arrays for 16 hours show marked light-depressed reporter gene transcription, which indicates that under different blue light exposure conditions, there was hardly any light-induced reporter gene transcriptional activity. But when in the dark environment (packaged with three layers of aluminum foil), our systems showed extremely high GFP expression (Figure 2).
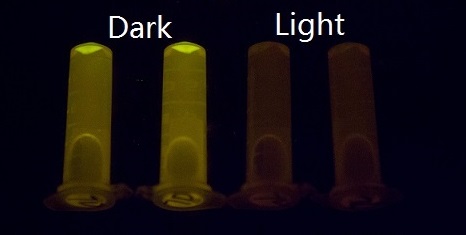
Figure 2. GFP expression was repressed by Blue LED light and presented normally in the dark.
There are a few plausible and conceivable reasons for why our system possesses such high sensitivity that can sense unbelievably weak light intensity. Firstly, it includes the unusually stable, photo-activated state of VVD, which causes the system to be extremely sensitive to light. Secondly, without relying on the addition of chromophores, which might impede the growth of cells, our system can directly regulate gene expression without trade-off.
2. Quantitative Result
Cells exposed to different light intensity expressing the Luminesensor showed manifest light-repressed reporter gene transcription. As shown in Figure 3, all of the cells with dissimilar attenuators showed incredible repression efficiency.

Figure 3. luminance attenuation using different attenuators could also be sensed by the Luminesensor, which is much dimmer than natural light.
It proves that once the cells are exposed to natural light, the transcription of reporter genes will be strongly repressed, although still present as a dose response. Besides, in the negative control group, which was entirely in the dark state, the expression of GFP ran up to a high degree of 50,000. As a matter of fact, as you can see later in "Results of light communication between cells", when we serially diluted light-emitting cells which express bacterial luciferase, the cells expressing the Luminesensor present significant dose response. Taking everything into account, our Luminesensor does possess high sensitivity across several orders of magnitude.
Orthogonality Test
Two sections of testing expriments were carried out. GFP was selected as a reporter and was fused downstream to two promoters (psulA408 and precA408) controlled by the Luminesensor with the 408 mutation and to two promoters (psulA and precA) controlled by the Luminesensor without the mutation. GFP expression is expected to have a negative relation with repression activity. To be more specific, higher level of green fluorescent indicates weaker repression effect; lower expression of GFP stands for stronger repression.
1. Experimental Design(Figure 4)
To prove the orthogonality, data and facts to prove that LexA408-VVD and the endogenous LexA work totally independently are needed. Considering the practical efficiency, two points of evidence were to be collected:
- 1. Promoters psulA and precA are repressed in wild-type Ecoli strains while promoters psulA408 and precA408 are not blocked in wild-type strains;
- 2. LexA408-VVD-Luminesensor efficiently represses its target under blue illumination, while it does not repress targets in total darkness.
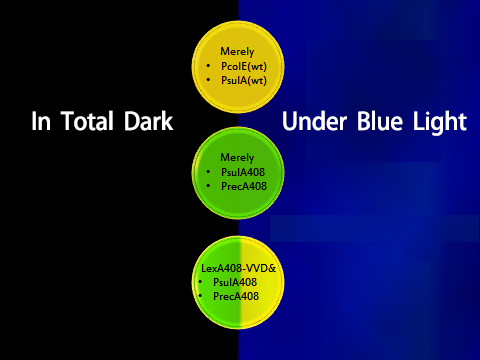
Figure 4. Shows observable color of plates where cell cultures growing under totally dark or light conditions. Different colors are used to indicate three levels of GFP expression.
2. Detailed Methods
Experiments were carried out in parallel -- taking photos of the plates as visual evidence and measuring GFP to quantify the effect. Detailed protocols are as follows:
- 1. Visible display:
-
- Inoculate a single colony into 1.5ml centifuge tube, shaking cultivate cells at 250 rpm/min for 2-3 h at 30℃, then the cell culture is ready for streaking;
- Streak cell culture onto respective plates;
- Incubate in total darkness and under light state at 30℃ for 1 or 2 days;
- Take photos of correct plates;
- 2. Data collection:
-
- Inoculate a proper single colony into liquid media;
- Cultivate cells in a shaking state until platform stage at 30 ℃;
- Dilute cell cultures with selective media at a ratio of 1:500;
- Cultivate cells in a shaking state until the stationary stage at 30 ℃ in complete darkness or under blue light;
- Harvest cells by centrifugation, then re-suspend pellets in PBS;
- Measure GFP expression with 96-well-microplate reader;
3. Results
Below is a collage of our plates. The plates are placed in groups at 30℃ in either total darkness or under blue-light illumination. Visual results fit well with Figure 4. Visual appearance of the plates were positive evidence for us.
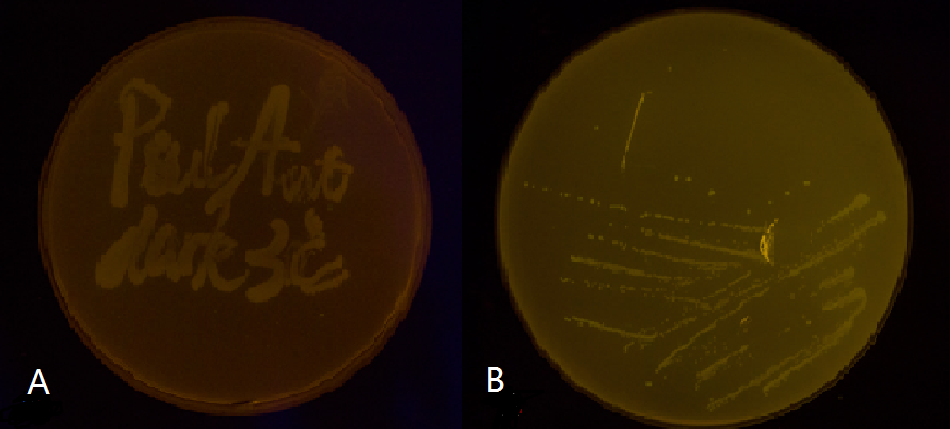
Figure 5(1). Plate streaked with wild type E.coli transformed with wild type Luminesensor and GFP fused to LexA responsive promoters, and cultivated in the dark (A), or under light (B). Evidently there is no GFP expression, indicating that endogenous LexA proteins are capable of fully repress the expression of our wild type reporter gene, rendering our Luminesensor system no longer light-controllable.
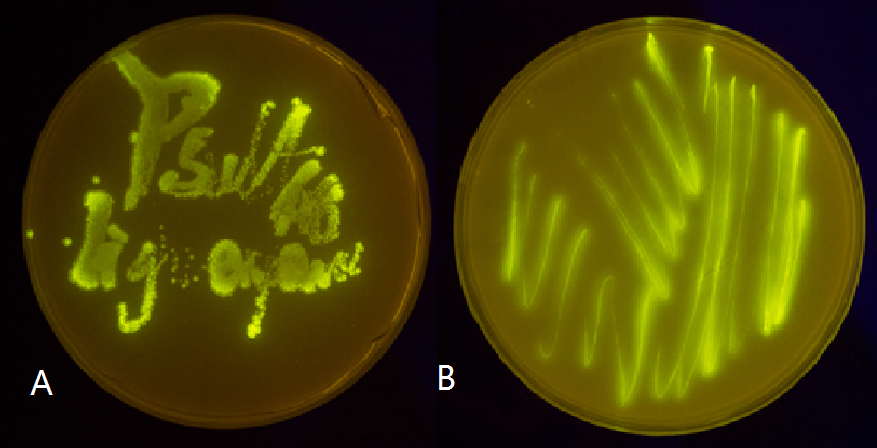
Figure 5(2). Plate streaked with wild type E.coli transformed with GFP gene fused to the downstream of psulA408 (A) and precA408 (B). Evidently GFP is perfectly expressed, indicating endogenous LexA proteins are unable to bind to psulA408 and precA408, which suggested that the 408 promoter system is orthogonal to the endogenous SOS system.
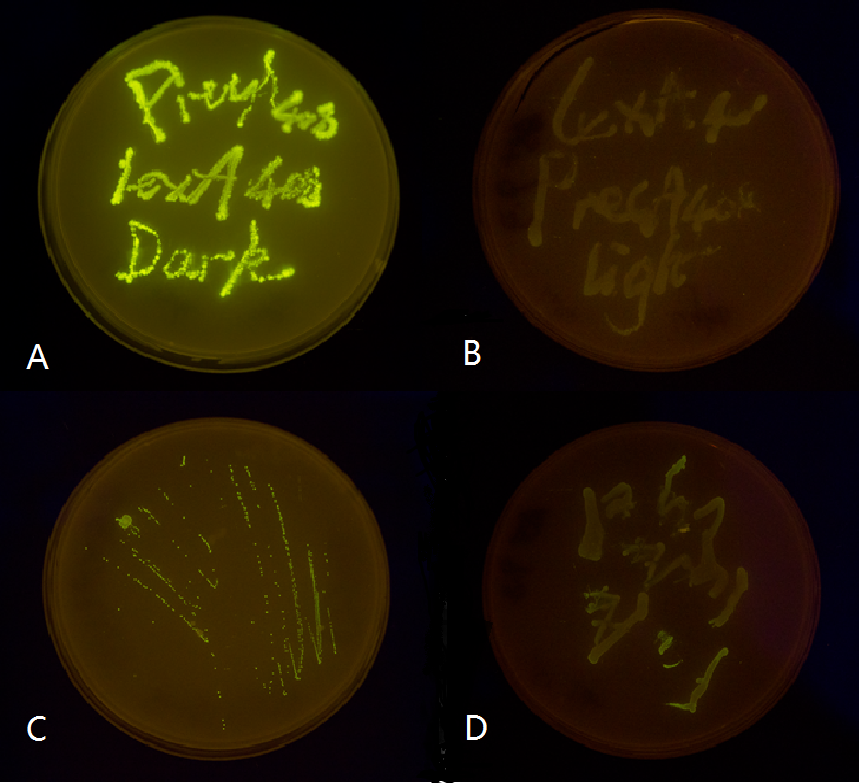
Figure 5(3). Plate streaked with wild type E.coli transformed with mutated luminesensor and precA408-GFP or psulA408-GFP reporter gene and cultivated in the dark or under light.(A: luminesensor + precA408-GFP, cultivated in the dark. B: luminesensor + precA408-GFP, cultivated under light. C: luminesensor + psulA408-GFP, cultivated in the dark. D: luminesensor + psulA408-GFP, cultivated under light.) Just as we predicted, the reporter gene is expressed in the dark and supressed by our luminesensor under light. This clearly proved that our 408 system is orthogonal to bacteria SOS system while retaining light-controllability.
Apart from those photos, more accurate results were obtained with Fluorescent Microplate Reader. Below are histograms based on our experimmental data. They clarify how fluoresence changes with lighting condition in different strains(Figure 6).
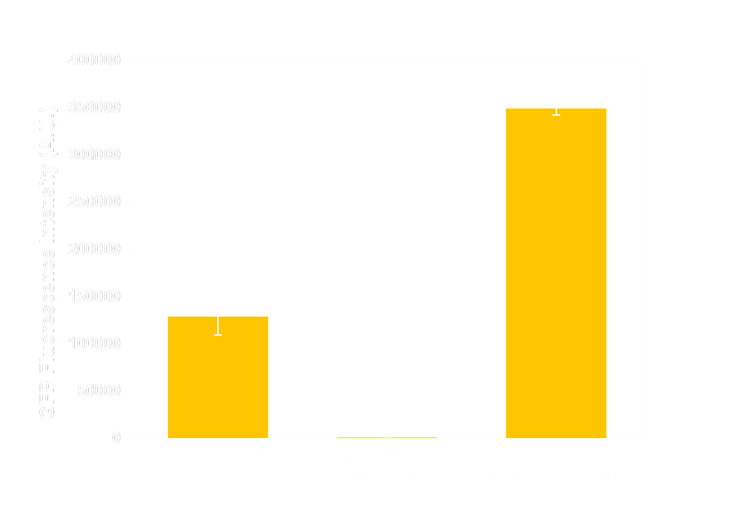
Figure 6(1). GFP expression level under control of precA408 promoter. Column 1 shows expression of precA-GFP reporter gene when transformed into wild-type E.coli without luminesensor. The high expression level indicates that the 408 reporter system is not interfered by endogenous LexA protein. The column 2 and column 3 are expression level of precA-GFP reporter gene co-transformed with our 408 form luminesensor into wild-type E.coli, and cultivated under light (column 2), or under light (column 3). The low expression level under light and hign expression level in the dark indicate that our 408 luminesensor system is totally light-controllable, and possessing a high dynamic range.
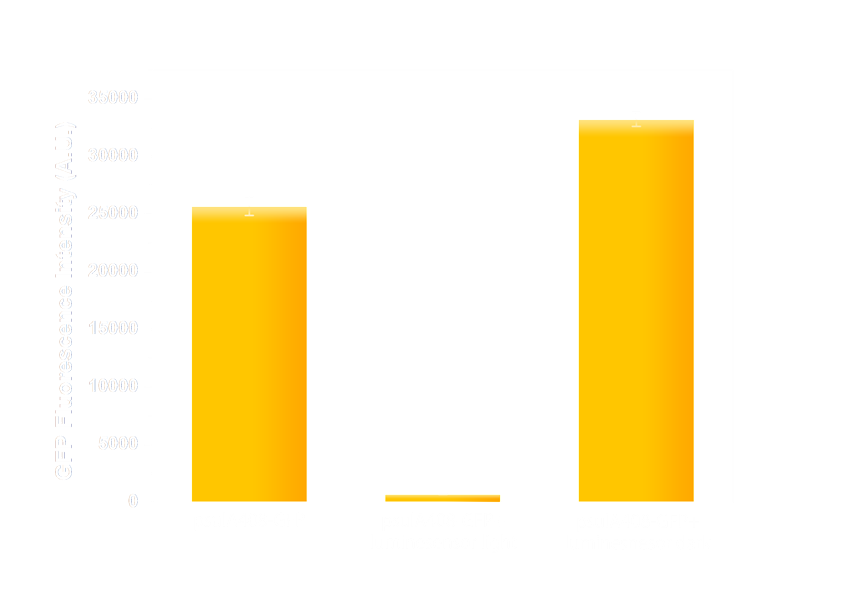
Figure 6(2). GFP expression level under control of psulA408 promoter. Every column shows data acquired under conditions same as its conterpart in Figure 7(1). The only difference is that the precA408 promoter has been replaced by psulA408 promoter.
It repeatedly turned out that:
1. reporters under wild-type promoters (psulA and precA) hardly expressed in either illuminated or dark state; their colonies are yellow. In contrast, GFPs fused to promoters psulA408 and precA408 were not blocked in strains, thus they looks bright green.
2. BL21 (DE3) strains expressing both the Luminesensor and its reporters shifted swiftly and sensitively from one states to the other, giving high definition output including lighting condition.
To sum up, LexA408-VVD-Luminesensor works independently of host genetic context.
Reference
- 1. Wang, X., Chen, X. & Yang, Y.(2012). spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods, 9: 266: 269
- 2. Farrell, C.M., Grossman, A.D., and Sauer., R.T.(2005). Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol., 57: 1750: 61
- 3. Roche, E.D., and Sauer., R.T.(2001). Identification of Endogenous SsrA-tagged Proteins Reveals Tagging at Positions Corresponding to Stop Codons. J. Biol. Chem., 276: 28509: 28515
- 4. Dimitrova,D., et al.(1997). A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli.Mol. Gen. Genet., 257: 205: 212
- 5. Voigt., C.A.(2006). Genetic parts to program bacteria. Curr. Opin. Biotechnol., 17: 548: 557
- 6. Thliveris, A.T., Mount., D.W.(1992). Genetic identification of the DNA binding domain of Escherichia coli LexA protein. Proc. Natl. Acad. Sci. USA, 89 (10): 4500: 4504
 "
"














