Team:SJTU-BioX-Shanghai/Project/project1.2
From 2012.igem.org
AleAlejandro (Talk | contribs) (→Fluorescence Complementation Assay) |
AleAlejandro (Talk | contribs) (→Construction Details) |
||
| (8 intermediate revisions not shown) | |||
| Line 63: | Line 63: | ||
Recruiting interacting protein domain and ligand on fusion membrane protein could help organize each component in order. | Recruiting interacting protein domain and ligand on fusion membrane protein could help organize each component in order. | ||
| - | [[File:12SJTU_constructionsketch.jpg|thumb| | + | [[File:12SJTU_constructionsketch.jpg|thumb|700px|center|''Fig.4 :''The construction sketch of fusion membrane protein]] |
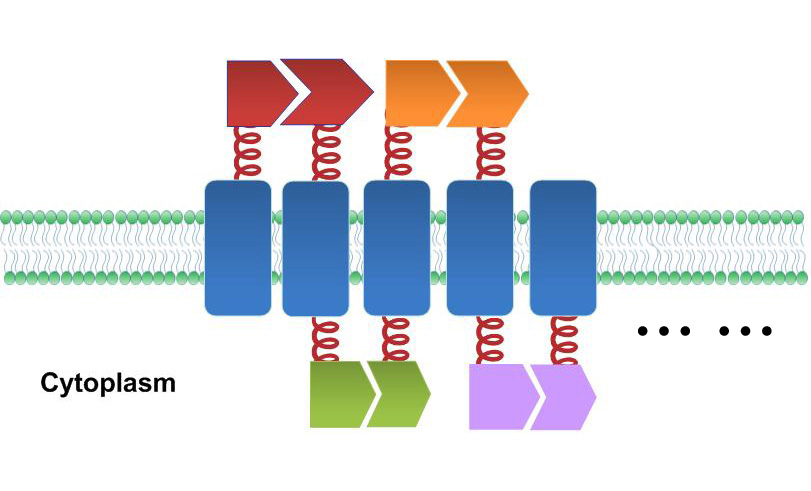
| - | [[File:12SJTU_membrane accelerator sketch.jpg|thumb|600px|center|Sketch of ''Membrane Accelerator''. Membrane proteins are organized through interacting proteins. Each pair of interacting proteins is shown in the same color]] | + | [[File:12SJTU_membrane accelerator sketch.jpg|thumb|600px|center|''Fig.5 :''Sketch of ''Membrane Accelerator''. Membrane proteins are organized through interacting proteins. Each pair of interacting proteins is shown in the same color]] |
==Fluorescence Complementation Assay== | ==Fluorescence Complementation Assay== | ||
| Line 75: | Line 75: | ||
EGFP was split into two halves, named 1EGFP and 2EGFP respectively. If there is interaction between two proteins which were fused with 1EGFP and 2EGFP, it is expected that fluorescence should be observed. Otherwise, no fluorescence could be observed. | EGFP was split into two halves, named 1EGFP and 2EGFP respectively. If there is interaction between two proteins which were fused with 1EGFP and 2EGFP, it is expected that fluorescence should be observed. Otherwise, no fluorescence could be observed. | ||
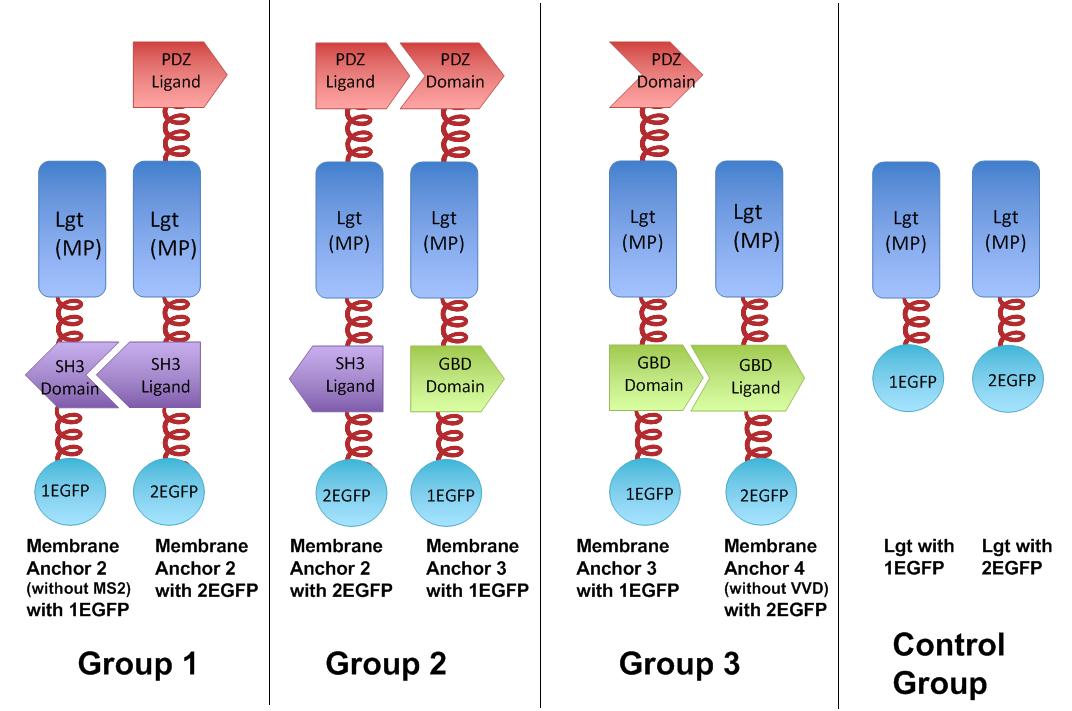
| - | The membrane proteins, which we call Membrane Anchors, are registered as [http://partsregistry.org/Part: | + | The membrane proteins, which we call Membrane Anchors, are registered as [http://partsregistry.org/Part:BBa_K771002 Part:BBa_K771002], [http://partsregistry.org/Part:BBa_K771003 Part:BBa_K771003], [http://partsregistry.org/Part:BBa_K771004 Part:BBa_K771004],[http://partsregistry.org/Part:BBa_K771005 Part:BBa_K771005], named Membrane Anchor 2 without MS2, Membrane Anchor 2,Membrane Anchor 3, Membrane Anchor 4 without VVD respectively. |
| - | [[Image:12SJTU、Dimertest.jpg|thumb|700px|center|''Fig. | + | [[Image:12SJTU、Dimertest.jpg|thumb|700px|center|''Fig.6'' :Group 1, 2, 3 are designed to test the interaction of SH3domain & ligand, PDZdomain & ligand and GBDdomain & ligand respectively. Proteins within each group were coexpressed in ''E.coli''. If there exists interaction within membrane proteins of each group, we expected to observe green fluorescence in ''E.coli''. Control group is expected to show much weaker fluorescence.]] |
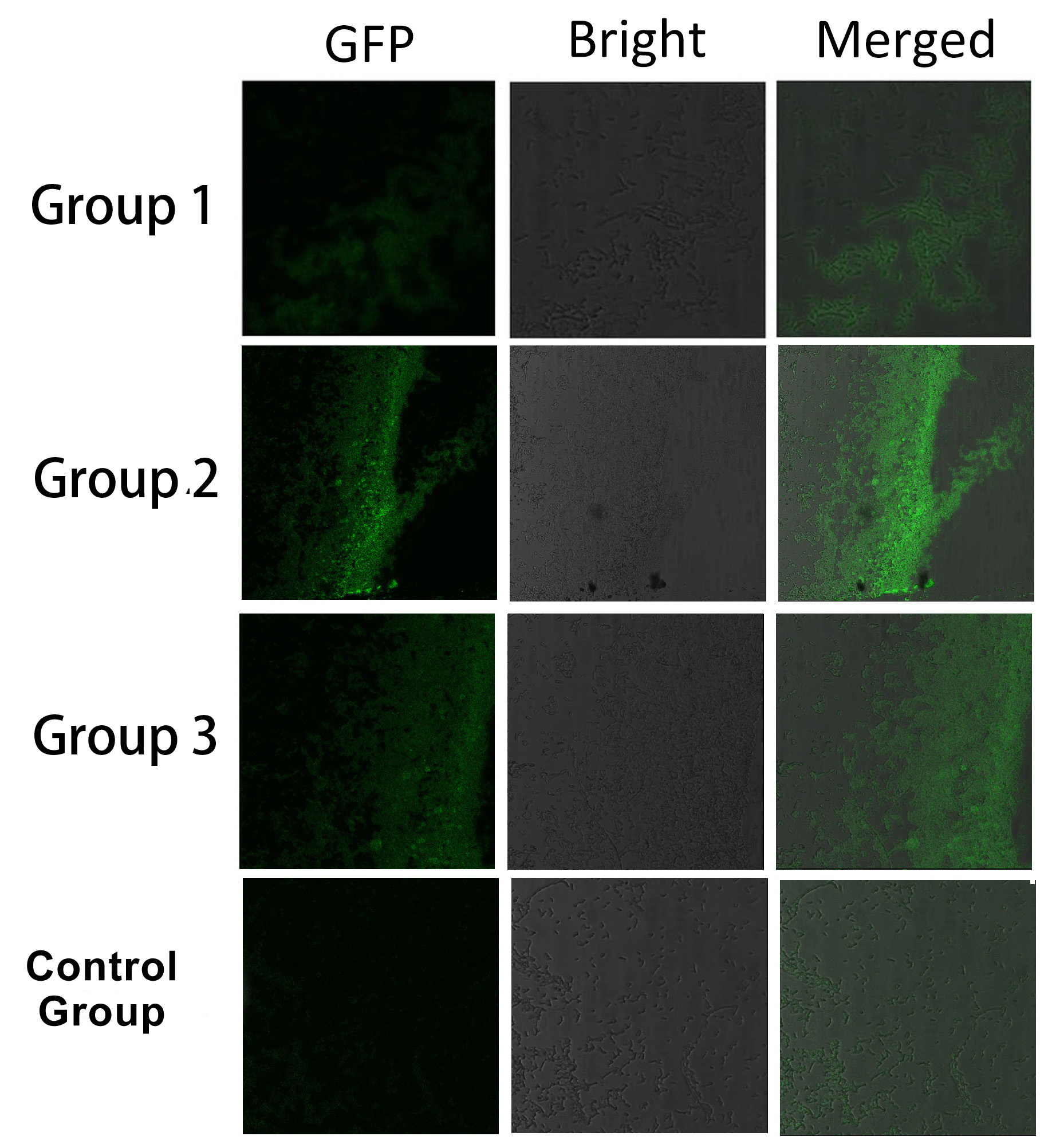
Proteins within each group are coexpressed in ''E.coli''. L-arabinose is not added into culture media until OD value of bacteria culture reaches 0.6. Bacteria are induced at L-Arabinose concentration of 0.2% for 6 hours. Bacteria samples are smeared onto glass slide and observed under laser confocal microscope. | Proteins within each group are coexpressed in ''E.coli''. L-arabinose is not added into culture media until OD value of bacteria culture reaches 0.6. Bacteria are induced at L-Arabinose concentration of 0.2% for 6 hours. Bacteria samples are smeared onto glass slide and observed under laser confocal microscope. | ||
| - | [[Image:12SJTU_SplitEGFP.jpg|thumb|700px|center|''Fig. | + | [[Image:12SJTU_SplitEGFP.jpg|thumb|700px|center|''Fig.7'' :Bacteria in Group 1, 2 and 3 all show significantly stronger fluorescence intensity than Control Group. So we can conclude that through recruiting interacting domain and ligand, we can assemble proteins into a complex.]] |
It is proved that membrane proteins with interacting proteins could interact and dimerize with each other. Thus, by recruiting ''E.coli'' native membrane protein Lgt, interacting proteins (SH3, PDZ, GBD) and downstream enzymes, we can easily build a ''Membrane Accelerator''. | It is proved that membrane proteins with interacting proteins could interact and dimerize with each other. Thus, by recruiting ''E.coli'' native membrane protein Lgt, interacting proteins (SH3, PDZ, GBD) and downstream enzymes, we can easily build a ''Membrane Accelerator''. | ||
| Line 87: | Line 87: | ||
==Application== | ==Application== | ||
| - | ''Membrane Accelerator'' could be easily put into use by '''attaching functional enzymes to | + | ''Membrane Accelerator'' could be easily put into use by '''attaching functional enzymes to corresponding membrane anchors'''. |
To verify the superiority of membrane accelerator system, we recruited [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project2.2 Fatty Acids Biosynthetic pathway], and proved that biosynthesis rate can be increased sharply with ''Membrane Accelerator''. | To verify the superiority of membrane accelerator system, we recruited [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project2.2 Fatty Acids Biosynthetic pathway], and proved that biosynthesis rate can be increased sharply with ''Membrane Accelerator''. | ||
Latest revision as of 02:28, 26 October 2012
| ||||||||||||||
|
 "
"