Team:LMU-Munich/Application
From 2012.igem.org
(Difference between revisions)
Franzi.Duerr (Talk | contribs) |
Franzi.Duerr (Talk | contribs) |
||
| (3 intermediate revisions not shown) | |||
| Line 34: | Line 34: | ||
{| width="100%" style="text-align:center;"| | {| width="100%" style="text-align:center;"| | ||
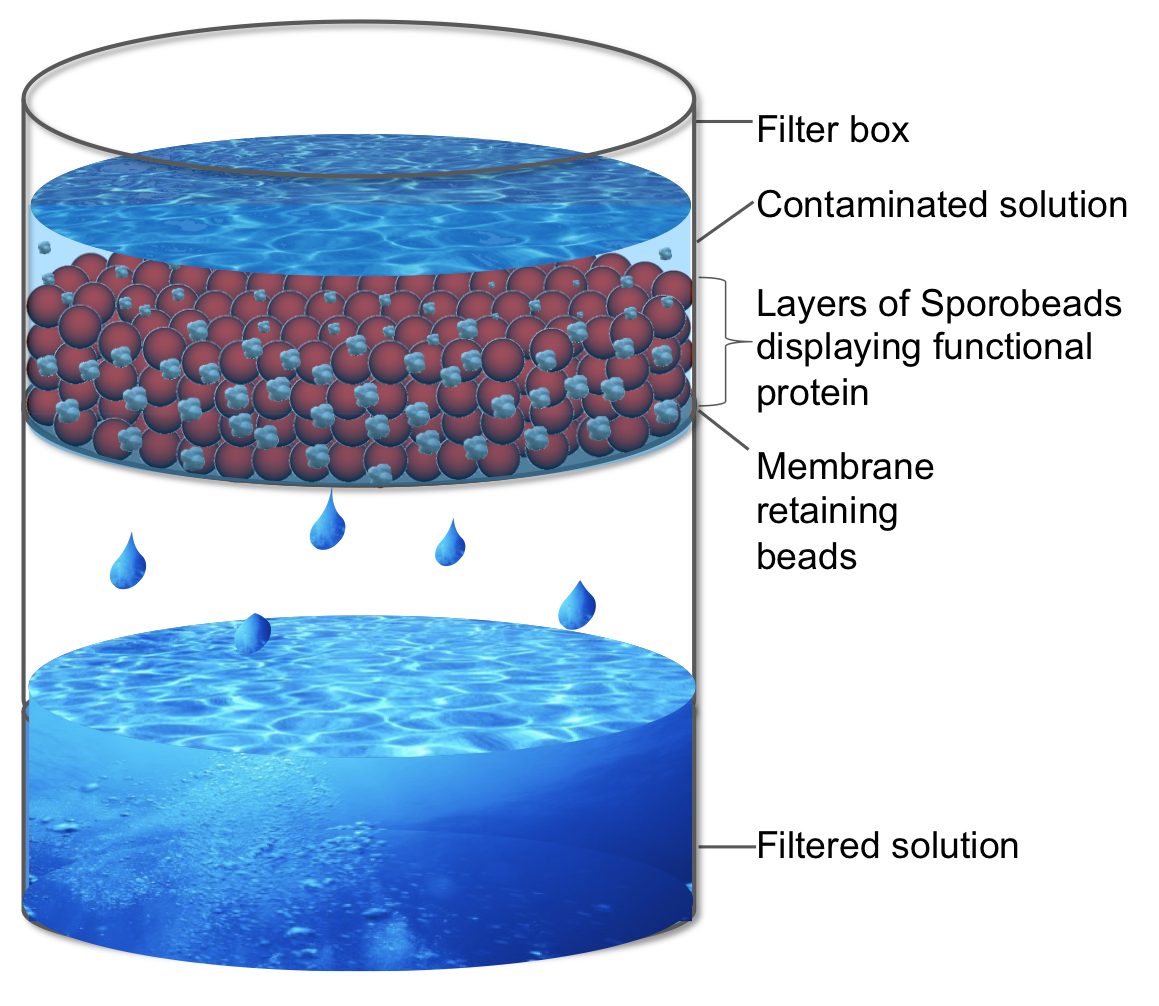
|<p align="justify">Further information for the first possible application for our '''Sporo'''beads.</p> | |<p align="justify">Further information for the first possible application for our '''Sporo'''beads.</p> | ||
| - | |[[File: | + | |[[File:LMU Sporofilter.png|right|150px|link=Team:LMU-Munich/Application/Filtration]] |
|- | |- | ||
! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Filtration]] | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Filtration]] | ||
| Line 44: | Line 44: | ||
{| width="100%" style="text-align:center;"| | {| width="100%" style="text-align:center;"| | ||
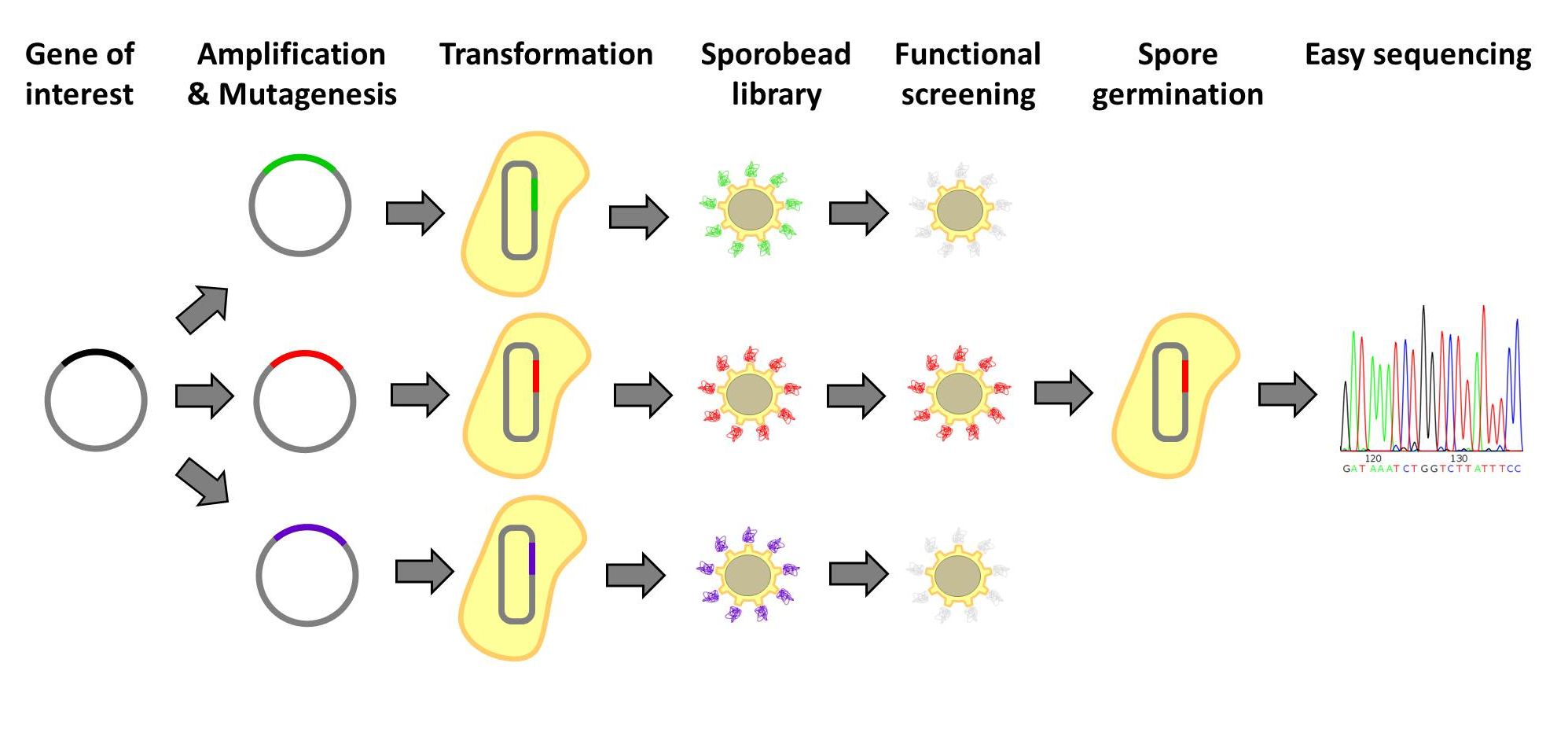
|<p align="justify">Further information for the possible application of protein screening.</p> | |<p align="justify">Further information for the possible application of protein screening.</p> | ||
| - | |[[File: | + | |[[File:protein_libraries.jpg|right|150px|link=Team:LMU-Munich/Application/Protein Screening]] |
|- | |- | ||
! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Protein Screening]] | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Protein Screening]] | ||
| Line 53: | Line 53: | ||
====Further Applications==== | ====Further Applications==== | ||
{| width="100%" style="text-align:center;"| | {| width="100%" style="text-align:center;"| | ||
| - | |<p align="justify"> | + | |<p align="justify">More information for further possible application with our diverse '''Sporo'''beads.</p> |
| - | |[[File: | + | |[[File:LMU Sporo diversity.png|right|150px|link=Team:LMU-Munich/Application/Further Applications]] |
|- | |- | ||
! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Further Applications]] | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Further Applications]] | ||
|} | |} | ||
</div> | </div> | ||
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
<div class="box"> | <div class="box"> | ||
====Project Navigation==== | ====Project Navigation==== | ||
Latest revision as of 23:24, 25 October 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


What are the potential advantages of using B. subtilis spores as functional bioparticals?
They are:
| Very stable even under severe environmental conditions | |
| Easy and cheap to produce in large quantities (check out the link to this commercial [http://www.alibaba.com/product-free/118791994/Bacillus_subtilis_spores_HU58_100_pure.html offer]) | |
| Part of human nutrition in terms of [http://en.wikipedia.org/wiki/Bacillus_subtilis_R0179 food supplements] and [http://www.fda.gov/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/default.htm generally regarded as safe] | |
| Moreover, our Sporobeads are: | |
| Unable to proliferate and therefore most likely not treated as genetically modified organisms by the law | |
Our Sporobeads offer a wide variety of applications within the laboratory, and around the world. Sporobeads can be used for filtration and protein screening.
 "
"