Team:LMU-Munich/Bacillus BioBricks/Reporters
From 2012.igem.org
(Created page with "<!-- Include the next line at the beginning of every page --> {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_Photo1.jpg}} [[File:NEXT.png|right|80px|link=Team:LM...") |
|||
| Line 3: | Line 3: | ||
| + | ==''Bacillus'' Reporters [[File:LMU Reporter.png|50px]]== | ||
| + | <p align="justify"> | ||
| + | We designed and codon-optimized a set of reporters that are commonly used in ''B. subtilis''. All reporters have a modified iGEM Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]) pre- and suffix for assembly of in-frame fusion proteins. Our prefix also includes the ''B. subitlis'' optimized ribosome binding site. </p> | ||
| + | Find out more about the design of our [https://static.igem.org/mediawiki/2012/b/b9/LMU-Munich_2012_Our_Freiburg_standard_FusionPrefix.pdf prefix with ribosome binding site]. | ||
| + | prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC | ||
| + | suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT | ||
| + | *'''GFP''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823039 BioBrick:BBa_K823039]) | ||
| - | [[File:NEXT.png|right|80px|link=Team:LMU-Munich/Bacillus_BioBricks]] [[File:BACK.png|left|80px|link=Team:LMU-Munich/Bacillus_BioBricks | + | |
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:WT-B53 fluorescence.png|300px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="90%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
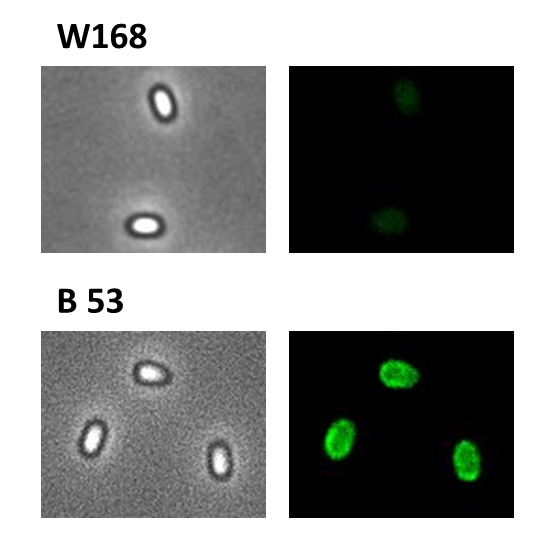
| + | <font color="#000000"; size="2"><p align="justify">Fig. 5: Fluorescence of wild type spores and B53-'''Sporo'''beads (containing the P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator construct)</p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | <p align="justify">We took a ''gfp'' derivate of the ''Bacillus subtilis'' plasmid pGFPamy and added the BioBrick compatible pre- and suffix of the Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]). The functionality of this BioBrick verified by creat e.g. our [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''beads].</p> | ||
| + | |||
| + | |||
| + | *'''mKate2''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 BioBrick:BBa_K823029]) | ||
| + | |||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:MKate Pellet_ii.JPG|400px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
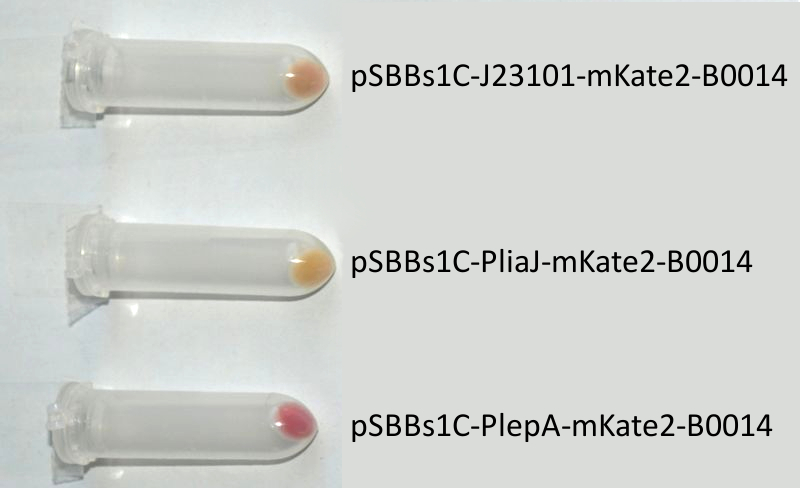
| + | <font color="#000000"; size="2"><p align="justify">'''''mKate2'' fused to the terminator B0014 under the control of the Anderson promoter J23101 (up), P<sub>''liaI''</sub> (middle) and P<sub>''lepA''</sub> (down) in pSB<sub>''Bs''</sub>1C.''' Pellets are ''Escherichia coli'' cells which contain the plasmid with the insert labelled to the right.</p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | <br> | ||
| + | |||
| + | <p align="justify">We codon-optimized the sequence of this monomeric far-red fluorescence protein for the use in ''B. subtilis'' and synthesized it (by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html GeneArt]) with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation, this reporter was successfully combined with the promoters P<sub>''liaI''</sub>, P<sub>''lepA''</sub> and the Anderson promoter J23101 in the empty ''Bacillus'' vector pSB<sub>''Bs''</sub>1C from our '''''Bacillus''B'''io'''B'''rick'''B'''ox. At the moment we already have the right construct integrated into the chromosome of ''B. subtilis''. Unfortunately, we have not yet measured this reporter.</p> | ||
| + | |||
| + | Reference: [http://www.evrogen.com/products/mKate2/mKate2.shtml mkate2] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:NEXT.png|right|80px|link=Team:LMU-Munich/Bacillus_BioBricks/Tags]] [[File:BACK.png|left|80px|link=Team:LMU-Munich/Bacillus_BioBricks]] | ||
Revision as of 20:27, 24 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Bacillus Reporters 
We designed and codon-optimized a set of reporters that are commonly used in B. subtilis. All reporters have a modified iGEM Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]) pre- and suffix for assembly of in-frame fusion proteins. Our prefix also includes the B. subitlis optimized ribosome binding site.
Find out more about the design of our prefix with ribosome binding site.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
- GFP ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823039 BioBrick:BBa_K823039])
|
We took a gfp derivate of the Bacillus subtilis plasmid pGFPamy and added the BioBrick compatible pre- and suffix of the Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]). The functionality of this BioBrick verified by creat e.g. our Sporobeads.
- mKate2 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 BioBrick:BBa_K823029])
|
We codon-optimized the sequence of this monomeric far-red fluorescence protein for the use in B. subtilis and synthesized it (by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html GeneArt]) with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation, this reporter was successfully combined with the promoters PliaI, PlepA and the Anderson promoter J23101 in the empty Bacillus vector pSBBs1C from our BacillusBioBrickBox. At the moment we already have the right construct integrated into the chromosome of B. subtilis. Unfortunately, we have not yet measured this reporter.
Reference: [http://www.evrogen.com/products/mKate2/mKate2.shtml mkate2]
 "
"