Team:Minnesota/Project/Caffeine Yeast
From 2012.igem.org
(Difference between revisions)
m |
|||
| (17 intermediate revisions not shown) | |||
| Line 132: | Line 132: | ||
<div id="MainBoxContent" style="background-color:white;"> | <div id="MainBoxContent" style="background-color:white;"> | ||
| - | <div style="position:absolute;top:0px;margin-left:50px; width:600px; height:450px; overflow-y:scroll; text-align:left;"> <!---START HERE---> | + | <div style="position:absolute;top:0px;margin-left:50px; width:600px; height:450px; overflow-y:scroll; text-align:left; overflow-x:hidden;"> <!---START HERE---> |
<h1>Engineering Caffeine Biosynthesis in Baker’s Yeast</h1><br> | <h1>Engineering Caffeine Biosynthesis in Baker’s Yeast</h1><br> | ||
| Line 144: | Line 144: | ||
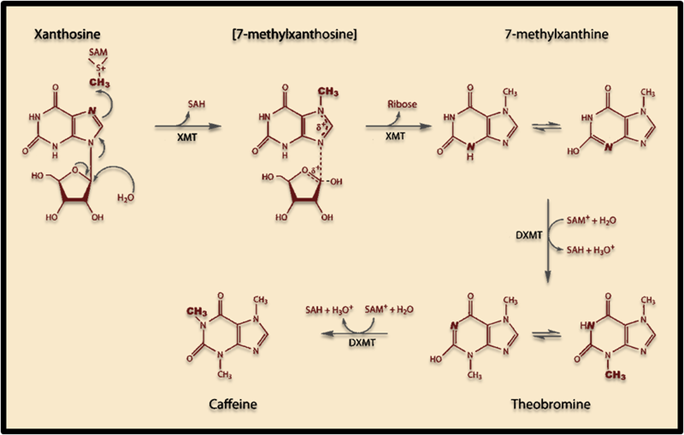
<p>The induction of this novel pathway in <i>S. cerevisiae</i> requires two additional enzymes from<i> C. canephora</i>: XMT1 and DXMT1. <i>S. cerevisiae</i> will produce the required metabolic precursors to Xanthosine, where after the XMT1 enzyme from <i>C. canephor</i>a will use the substrate to synthesize 7-methylxanthosine, then 7-Methylxanthine. DXMT1 will then convert it to Theobromine and finally caffeine. </p> | <p>The induction of this novel pathway in <i>S. cerevisiae</i> requires two additional enzymes from<i> C. canephora</i>: XMT1 and DXMT1. <i>S. cerevisiae</i> will produce the required metabolic precursors to Xanthosine, where after the XMT1 enzyme from <i>C. canephor</i>a will use the substrate to synthesize 7-methylxanthosine, then 7-Methylxanthine. DXMT1 will then convert it to Theobromine and finally caffeine. </p> | ||
| - | + | ||
| + | <img style="width:500px;" src="http://i1158.photobucket.com/albums/p607/iGEM_MN/caffeine_synth_resize.png"> | ||
| + | |||
<p><b>Figure 1. Scheme showing proposed caffeine biosynthetic pathway in S. cerevisiae. </b></p> | <p><b>Figure 1. Scheme showing proposed caffeine biosynthetic pathway in S. cerevisiae. </b></p> | ||
| Line 150: | Line 152: | ||
After settling on the two genes, the sequences were taken from NCBI and finalized for synthetic synthesis (XMT1 accession number DQ422954 and DXMT1 accession number DQ422955). A program was designed to optimize these plant genes for yeast use. Because of the similarity of the two genes (~88% similarity) the program was also designed to identify regions of homology between the two genes and adjust the codon usage such that homologous recombination will not occur at the nucleotide level while the overall protein sequence would be retained. The codons were chosen in descending order of complementary tRNA abundance. Any BioBrick cut sites found within the gene sequences were also removed.</p> | After settling on the two genes, the sequences were taken from NCBI and finalized for synthetic synthesis (XMT1 accession number DQ422954 and DXMT1 accession number DQ422955). A program was designed to optimize these plant genes for yeast use. Because of the similarity of the two genes (~88% similarity) the program was also designed to identify regions of homology between the two genes and adjust the codon usage such that homologous recombination will not occur at the nucleotide level while the overall protein sequence would be retained. The codons were chosen in descending order of complementary tRNA abundance. Any BioBrick cut sites found within the gene sequences were also removed.</p> | ||
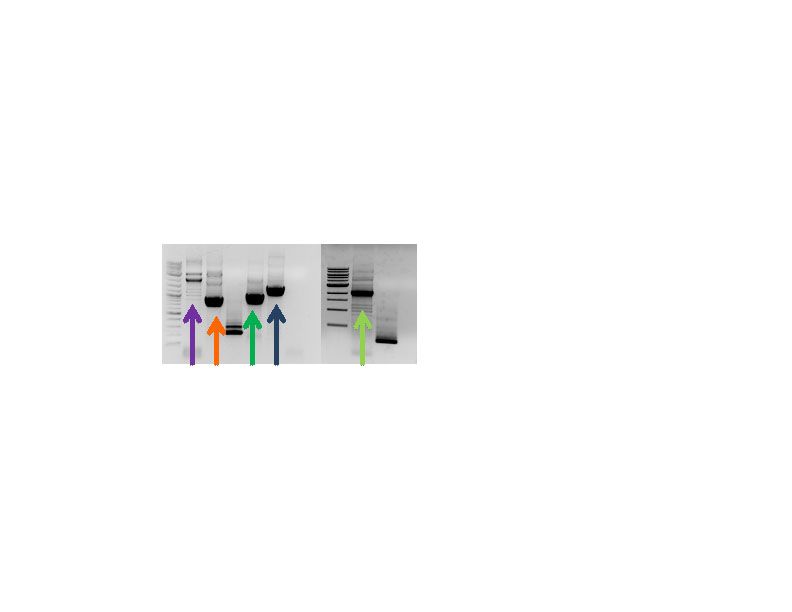
| - | <p>To synthesize the overall plasmid backbone, PCR primers were designed to amplify five desired fragments (with 25-30 bp overlap) from different plasmids, which could be joined by Gibson Assembly: | + | <p>To synthesize the overall plasmid backbone, PCR primers were designed to amplify five desired fragments (with 25-30 bp overlap) from different plasmids, which could be joined by Gibson Assembly:<br> |
| - | + | 1. BioBrick destination plasmid pSB1C3 containing the MCS and rep (pMB1).<br> | |
| - | 1. BioBrick destination plasmid pSB1C3 containing the MCS and rep (pMB1).<br> | + | 2. BioBrick destination plasmid pSB1C3 containing the Chloramphenical resistance gene. <br> |
| - | 2. BioBrick destination plasmid pSB1C3 containing the Chloramphenical resistance gene. <br> | + | 3. 2u Gibson Extract provided by the Schmidt-Dannert lab (University of Minnesota) containing the 2u ORI for yeast. <br> |
| - | 3. 2u Gibson Extract provided by the Schmidt-Dannert lab (University of Minnesota) containing the 2u ORI for yeast. <br> | + | 4. pkT127 provided by the Schmidt-Dannert lab containing G418 resistance genes and tTEF1.<br> |
| - | 4. pkT127 provided by the Schmidt-Dannert lab containing G418 resistance genes and tTEF1.<br> | + | 5. ATCC plasmid provided by the Schmidt-Dannert lab containing the ADH1 promoter to drive the G418 R gene.</p> |
| - | 5. ATCC plasmid provided by the Schmidt-Dannert lab containing the ADH1 promoter to drive the G418 R gene.</p> | + | |
| + | <p><b>Parts List</b><br> | ||
| + | BBa_K814000 dehydroquinate synthase (DHQS) generator<br> | ||
| + | BBa_K814001 ATP-grasp (ATPG) generator<br> | ||
| + | |||
| + | BBa_K814002 dehydroquinate O-methyltrasferase (O-MT) generator<br> | ||
| + | |||
| + | BBa_K814003 shinorine non-ribosomal peptide synthase (NRPS) generator<br> | ||
| + | |||
| + | BBa_K814004 mycosporine-glycine biosynthetic pathway<br> | ||
| + | |||
| + | BBa_K814005 shinorine biosynthetic pathway<br> | ||
| + | |||
| + | BBa_K814006 negative control for mycosporine-like amino acid biosynthesis<br> | ||
| + | |||
| + | BBa_K814007 ScyA (acetolactate synthase) generator<br> | ||
| + | |||
| + | BBa_K814008 ScyB (leucine dehydrogenase) generator<br> | ||
| + | |||
| + | BBa_K814009 ScyC generator<br> | ||
| + | |||
| + | BBa_K814010 partial scytonemin biosynthetic pathway, scyCB<br> | ||
| + | |||
| + | BBa_K814011 scytonemin biosynthetic pathway, scyBAC<br> | ||
| + | |||
| + | BBa_K814012 XMT1 protein generator<br> | ||
| + | |||
| + | BBa_K814013 DXMT1 protein generator | ||
| + | </p> | ||
| + | |||
| + | <img src="http://i1158.photobucket.com/albums/p607/iGEM_MN/colorarrowscopy.jpg"> | ||
| + | |||
| + | |||
| + | <p><b>Figure 2:</b> Pieces for Gibson assembly of our novel, shuttle backbone. The individual pieces relate to the described components enumerated above. 1. Purple arrow; 2. Orange arrow; 3. Green arrow; 4. Blue arrow; 5. Light green arrow.</p> | ||
| + | |||
| + | <img src="http://i1158.photobucket.com/albums/p607/iGEM_MN/cells.jpg | ||
| + | "> | ||
| + | |||
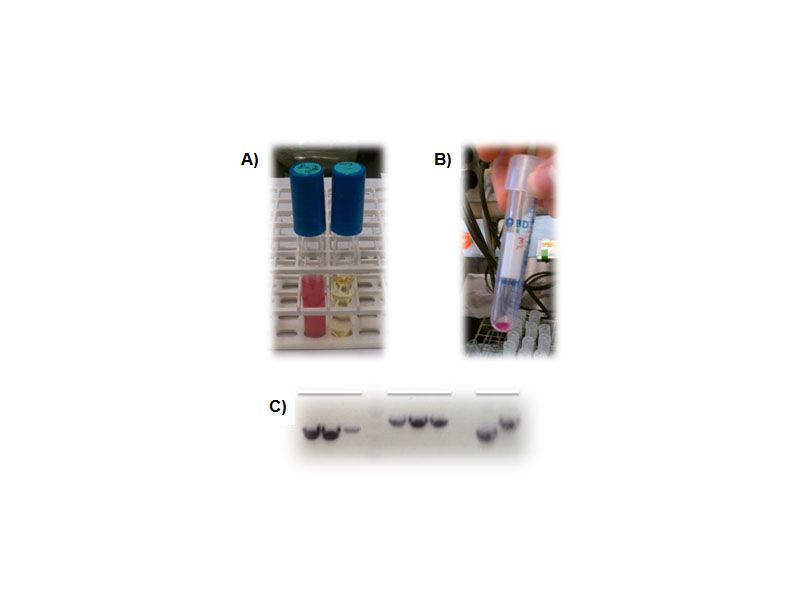
| + | <p><b>Figure 3. Creation of a shuttle vector designed for BioBrick components.</b> A), Overnight culture of the pSB1C3 shipping vector showing RFP expression (a blank LB tube is shown as a control on the right). B), RFP expression in the pGHMM2012 shuttle vector. Cell pellets clearly indicate expression of RFP from this plasmid. C), PCR screening for caffeine biosynthetic components into pGHMM2012. The panels from left to right are positive clones for Xmt, Dxmt and controls for each of these components.</p> | ||
| + | |||
| + | |||
| + | <img style="width:500px;" src="http://i1158.photobucket.com/albums/p607/iGEM_MN/growth_curve_resize.png"> | ||
| + | |||
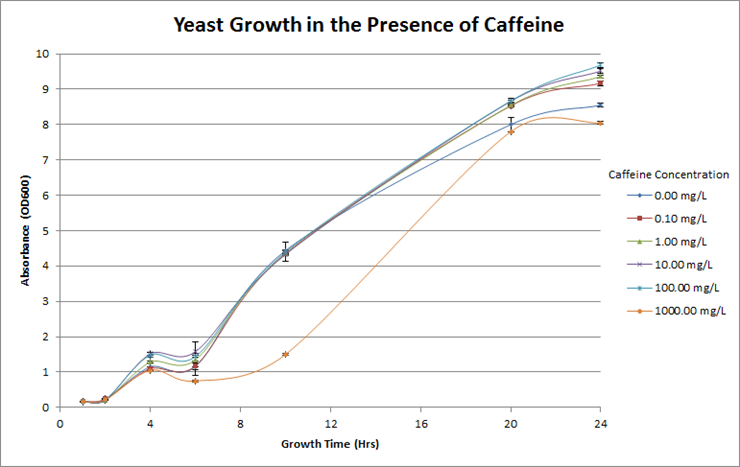
| + | <p><b>Figure 4. Investigation of caffeine toxicity in yeast. </b>The yeast were resilient in the presence of caffeine and there was no significant decrease in growth overtime at each different concentration. For reference, the concentration of caffeine in coffee is roughly 600mg/L.</p> | ||
| + | |||
| + | <img src="http://i1158.photobucket.com/albums/p607/iGEM_MN/hplccaffeine-1_resize.jpg"> | ||
| + | |||
| + | |||
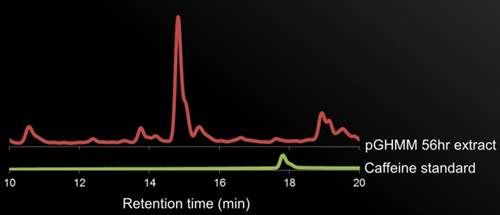
| + | <p><b>Figure 5. HPLC Data. </b>description.</p> | ||
| + | |||
| + | <p><b>Conclusions</b><br> | ||
| + | We were able to design and synthesize a novel yeast- E. coli hybrid plasmid which was optimized to prevent homologous recombination in our two synthetic genes: XMT1 and DXMT1. </p> | ||
| + | |||
| + | <p>If the HPLC results can confirm the presence of caffeine in the yeast cultures, we would confirm the presence of the two target proteins by SDS-PAGE, possibly even performing antibody detection if there is difficulty confirming the presence.</p> | ||
| + | |||
| + | <p>If the HPLC results cannot confirm the presence of caffeine, the first step would be to sequence our genes and align them with the sequences submitted to IDT for the gBlocks. We could also measure the production of the intermediates compared to the cultures to test whether caffeine synthesis is being arrested in the pathway.</p> | ||
| + | |||
<!---END--> | <!---END--> | ||
Latest revision as of 04:02, 4 October 2012

Like us on FB and follow us on Twitter!
 "
"