|
|
| Line 344: |
Line 344: |
| | <p><span style="color:#000000;"><em>CheB</em> removes the methyl groups that are attached to the gamma-glutamyl methyl ester residues located in the methyl-accepting chemotaxis protein (MCP). This allows for the bacteria to remain alert to the changes in the environment.</span></p> | | <p><span style="color:#000000;"><em>CheB</em> removes the methyl groups that are attached to the gamma-glutamyl methyl ester residues located in the methyl-accepting chemotaxis protein (MCP). This allows for the bacteria to remain alert to the changes in the environment.</span></p> |
| | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> | | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842007"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842007</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842007"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842007</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 361: |
Line 361: |
| | <p><span style="color:#000000;"><em>CheY</em> is a regulatory protein that reverses the direction of flagella rotation from counter-clockwise to clockwise. When the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant, <em>cheA</em> is activated and told to self-phosphorylate itself by hydrolyzing ATP. Once the <em>cheA</em> is phosphorylated, it transfers its phosphate group to <em>cheY</em>, therefore activating it. <em>CheY</em> then moves to the cytoplasmic side of the flagella apparatus and binds to it. When flagella is not bound to <em>cheY</em>, it rotates in a counter-clockwise direction, which allows the bacteria to move in a “forward” direction. Flagella that is bound to phosphorylated <em>cheY</em> switches its rotation to clockwise, which induces tumbling.</span></p> | | <p><span style="color:#000000;"><em>CheY</em> is a regulatory protein that reverses the direction of flagella rotation from counter-clockwise to clockwise. When the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant, <em>cheA</em> is activated and told to self-phosphorylate itself by hydrolyzing ATP. Once the <em>cheA</em> is phosphorylated, it transfers its phosphate group to <em>cheY</em>, therefore activating it. <em>CheY</em> then moves to the cytoplasmic side of the flagella apparatus and binds to it. When flagella is not bound to <em>cheY</em>, it rotates in a counter-clockwise direction, which allows the bacteria to move in a “forward” direction. Flagella that is bound to phosphorylated <em>cheY</em> switches its rotation to clockwise, which induces tumbling.</span></p> |
| | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> | | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842009"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842009</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842009"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842009</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 378: |
Line 378: |
| | <p><span style="color:#000000;"><em>CheZ</em> is a regulatory protein that constantly dephosphorylates <em>cheY</em> that is attached to a flagellum. The incessant dephosphorylation is what causes bacteria to run, stop, and tumble when in an environment that has a concentration gradient and remain responsive to any changes in chemical concentration.</span></p> | | <p><span style="color:#000000;"><em>CheZ</em> is a regulatory protein that constantly dephosphorylates <em>cheY</em> that is attached to a flagellum. The incessant dephosphorylation is what causes bacteria to run, stop, and tumble when in an environment that has a concentration gradient and remain responsive to any changes in chemical concentration.</span></p> |
| | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> | | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 395: |
Line 395: |
| | <p><span style="color:#000000;"><em>motA</em> is a structural protein for the stationary element of the flagella motor. The protein works in conjunction with <em>motB</em> to form this structure, which then in turn controls the flagella’s rotation.</span></p> | | <p><span style="color:#000000;"><em>motA</em> is a structural protein for the stationary element of the flagella motor. The protein works in conjunction with <em>motB</em> to form this structure, which then in turn controls the flagella’s rotation.</span></p> |
| | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> | | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 429: |
Line 429: |
| | <p><span style="color:#000000;">T7 serves as a promoter that is stimulated by IPTG. RBS is a sequence in DNA located upstream of the start codon. It affects the rate at which the open reading frame is translated. <em>CheZ</em> is a regulatory protein that constantly dephosphorylates <em>cheY</em> that is attached to a flagellum. The incessant dephosphorylation is what causes bacteria to run, stop, and tumble when in an environment that has a concentration gradient and remain responsive to any changes in chemical concentration. Together, <em>cheZ</em> is induced by the presence of IPTG.</span></p> | | <p><span style="color:#000000;">T7 serves as a promoter that is stimulated by IPTG. RBS is a sequence in DNA located upstream of the start codon. It affects the rate at which the open reading frame is translated. <em>CheZ</em> is a regulatory protein that constantly dephosphorylates <em>cheY</em> that is attached to a flagellum. The incessant dephosphorylation is what causes bacteria to run, stop, and tumble when in an environment that has a concentration gradient and remain responsive to any changes in chemical concentration. Together, <em>cheZ</em> is induced by the presence of IPTG.</span></p> |
| | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> | | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 446: |
Line 446: |
| | <p><span style="color:#000000;">T7 serves as a promoter that is stimulated by IPTG. RBS is a sequence in DNA located upstream of the start codon. It affects the rate at which the open reading frame is translated. <em>CheY</em> is regulatory protein that reverses the direction of flagella rotation from counter-clockwise to clockwise. When the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant, <em>cheA</em> is activated and told to self-phosphorylate itself by hydrolyzing ATP. Once the <em>cheA</em> is phosphorylated, it transfers its phosphate group to <em>cheY</em>, therefore activating it. <em>CheY</em> then moves to the cytoplasmic side of the flagella apparatus and binds to it. When flagella are not bound to <em>cheY</em>, it rotates in a counter-clockwise direction, which allows the bacteria to move in a “forward” direction. Flagella that are bound to phosphorylated <em>cheY</em> switches its rotation to clockwise, which induces tumbling.</span></p> | | <p><span style="color:#000000;">T7 serves as a promoter that is stimulated by IPTG. RBS is a sequence in DNA located upstream of the start codon. It affects the rate at which the open reading frame is translated. <em>CheY</em> is regulatory protein that reverses the direction of flagella rotation from counter-clockwise to clockwise. When the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant, <em>cheA</em> is activated and told to self-phosphorylate itself by hydrolyzing ATP. Once the <em>cheA</em> is phosphorylated, it transfers its phosphate group to <em>cheY</em>, therefore activating it. <em>CheY</em> then moves to the cytoplasmic side of the flagella apparatus and binds to it. When flagella are not bound to <em>cheY</em>, it rotates in a counter-clockwise direction, which allows the bacteria to move in a “forward” direction. Flagella that are bound to phosphorylated <em>cheY</em> switches its rotation to clockwise, which induces tumbling.</span></p> |
| | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> | | <p><span style="color:#000000;">Used for researcher control of signaling mechanisms that influence flagella function.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842015"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842015</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842015"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842015</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 463: |
Line 463: |
| | <p><span style="color:#000000;"><em>flgJ</em> transcribes a flagellum specific muramidase which hydrolyzes the peptidoglycan layer in the cell membrane. This creates a small hole in the side of the bacterium in which the flagellar rod is formed. It is most commonly believed that <em>flgJ</em> is exported via the flagellum specific exportation system, making it a class 3 flagella gene.</span></p> | | <p><span style="color:#000000;"><em>flgJ</em> transcribes a flagellum specific muramidase which hydrolyzes the peptidoglycan layer in the cell membrane. This creates a small hole in the side of the bacterium in which the flagellar rod is formed. It is most commonly believed that <em>flgJ</em> is exported via the flagellum specific exportation system, making it a class 3 flagella gene.</span></p> |
| | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> | | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 481: |
Line 481: |
| | <p><span style="color:#000000;">fliA is an alternate sigma factor for the class 3 flagella operons. When transcribed, it forms a component of RNA polymerase sigma 28.</span></p> | | <p><span style="color:#000000;">fliA is an alternate sigma factor for the class 3 flagella operons. When transcribed, it forms a component of RNA polymerase sigma 28.</span></p> |
| | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> | | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 500: |
Line 500: |
| | <p><span style="color:#000000;">FliH functions as part of the flagella specific export system and is necessary for flagellar growth and assembly. It is believed to be part of a two-component system consisting of fliH and fliL that hydrolyses ATP until construction of the flagella apparatus is completed.</span></p> | | <p><span style="color:#000000;">FliH functions as part of the flagella specific export system and is necessary for flagellar growth and assembly. It is believed to be part of a two-component system consisting of fliH and fliL that hydrolyses ATP until construction of the flagella apparatus is completed.</span></p> |
| | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> | | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842005"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842005</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842005"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842005</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 516: |
Line 516: |
| | <td valign="top" width="420"><span style="color:#000000;"><strong>Description and Function</strong></span></p> | | <td valign="top" width="420"><span style="color:#000000;"><strong>Description and Function</strong></span></p> |
| | <p><span style="color:#000000;">When transcribed, RBS-fliC-GFP generates the flagellum protein bonded with a green fluorescent protein. This structure would then be exported to the growing flagella apparatus and used in the construction of the filament. When strongly expressed, this structure would directly result in the flagella filament's containing GFP, allowing for the flagella to be detectable. Unfortunately we were unable to promote the sequence enough to test whether this construct actually functions in this manner.</span></p> | | <p><span style="color:#000000;">When transcribed, RBS-fliC-GFP generates the flagellum protein bonded with a green fluorescent protein. This structure would then be exported to the growing flagella apparatus and used in the construction of the filament. When strongly expressed, this structure would directly result in the flagella filament's containing GFP, allowing for the flagella to be detectable. Unfortunately we were unable to promote the sequence enough to test whether this construct actually functions in this manner.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 534: |
Line 534: |
| | <td valign="top" width="420"><span style="color:#000000;"><strong>Description and Function</strong></span></p> | | <td valign="top" width="420"><span style="color:#000000;"><strong>Description and Function</strong></span></p> |
| | <p><span style="color:#000000;">Class 2 flagella operon. When transcribed, flhB is one of three flagella genes responsible for the generation of proteins that export flagellin to the flagella apparatus. The flagellin is then directly used in the synthesis of the structure's basal body. Additionally, flhB directly transcribes the operon flhBAE.</span></p> | | <p><span style="color:#000000;">Class 2 flagella operon. When transcribed, flhB is one of three flagella genes responsible for the generation of proteins that export flagellin to the flagella apparatus. The flagellin is then directly used in the synthesis of the structure's basal body. Additionally, flhB directly transcribes the operon flhBAE.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842000"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842000</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842000"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842000</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| Line 553: |
Line 553: |
| | <p><span style="color:#000000;">flhD along with flhC form the master regulation operon for the synthesis of flagella in<em> E. coli. </em> They are responsible for the downstream transcription of the class 2 flagella genes. flhD is directly induced by several separate systems; among these is the two-component quorum sensing response system, qseB and qseC.</span></p> | | <p><span style="color:#000000;">flhD along with flhC form the master regulation operon for the synthesis of flagella in<em> E. coli. </em> They are responsible for the downstream transcription of the class 2 flagella genes. flhD is directly induced by several separate systems; among these is the two-component quorum sensing response system, qseB and qseC.</span></p> |
| | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> | | <p><span style="color:#000000;">Used for reconstitution of flagella apparatus in <em>E. coli</em> B strains.</span></p> |
| - | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842001"><span style="color:#000000;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842001</span></a></span></td> | + | <p><span style="color:#000000;"><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K842001"><span style="color:#0000FF;">http://partsregistry.org/wiki/index.php?title=Part:BBa_K842001</span></a></span></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
Our BioBricks
Below are the BioBrick parts our team created and submitted to the iGEM Registry.
Click images to enlarge. Featured BioBricks are highlighted in yellow.

 |
BBa_K842006
cheA |
Description and Function
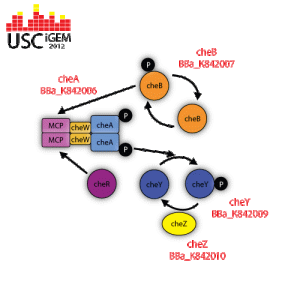
cheA is a regulatory protein that becomes activated when the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant. The activated cheA self-phosphorylates itself by hydrolyzing ATP in order to gain a phosphate group. cheA then donates its phosphate group to activate cheB or cheA.
Used for researcher control of signaling mechanisms that influence flagella function.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842005 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used with other chemotaxis genes to form a complete regulatory pathway that controls the response system in MCP and the direction of rotation in flagella. |
| Sources
Chemotaxis protein cheA – Escherichia coli (strain K12). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P07363 |
 |
BBa_K842007
cheB |
Description and Function
CheB removes the methyl groups that are attached to the gamma-glutamyl methyl ester residues located in the methyl-accepting chemotaxis protein (MCP). This allows for the bacteria to remain alert to the changes in the environment.
Used for researcher control of signaling mechanisms that influence flagella function.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842007 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used with other chemotaxis genes to form a complete regulatory pathway that controls the response system in MCP and the direction of rotation in flagella. |
| Sources
Chemotaxis response regulator protein-glutamate methylesterase – Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P04042 |
 |
BBa_K842009
cheY |
Description and Function
CheY is a regulatory protein that reverses the direction of flagella rotation from counter-clockwise to clockwise. When the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant, cheA is activated and told to self-phosphorylate itself by hydrolyzing ATP. Once the cheA is phosphorylated, it transfers its phosphate group to cheY, therefore activating it. CheY then moves to the cytoplasmic side of the flagella apparatus and binds to it. When flagella is not bound to cheY, it rotates in a counter-clockwise direction, which allows the bacteria to move in a “forward” direction. Flagella that is bound to phosphorylated cheY switches its rotation to clockwise, which induces tumbling.
Used for researcher control of signaling mechanisms that influence flagella function.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842009 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used with other chemotaxis genes to form a complete regulatory pathway that controls the response system in MCP and the direction of rotation in flagella. |
| Sources
Chemotaxis protein CheY – Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P0A2D5 |
 |
BBa_K842010
cheZ |
Description and Function
CheZ is a regulatory protein that constantly dephosphorylates cheY that is attached to a flagellum. The incessant dephosphorylation is what causes bacteria to run, stop, and tumble when in an environment that has a concentration gradient and remain responsive to any changes in chemical concentration.
Used for researcher control of signaling mechanisms that influence flagella function.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used with other chemotaxis genes to form a complete regulatory pathway that controls the response system in MCP and the direction of rotation in flagella. |
| Sources
Protein phosphatase CheZ – Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P07800 |
 |
BBa_K842011
motA |
Description and Function
motA is a structural protein for the stationary element of the flagella motor. The protein works in conjunction with motB to form this structure, which then in turn controls the flagella’s rotation.
Used for reconstitution of flagella apparatus in E. coli B strains.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. The protein works in conjunction with motB to form the stationary base of the flagella motor, which then in turn controls the flagella’s rotation. This part can be used in the complete recreation of an E. coli’s genetic flagellar pathway. |
| Sources
Motility protein A – Escherichia coli (strain K12). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P09348 |
 |
BBa_K842012
motB |
Description and Function
motB is a structural protein for the stationary element of the flagella motor. The protein works in conjunction with motA to form this structure, which then in turn controls the flagella’s rotation. It is also believed that motB is responsible for forming the structure that connects the flagella motor to the cell wall.
Used for reconstitution of flagella apparatus in E. coli B strains.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842010 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. The protein works in conjunction with motA to form the stationary base of the flagella motor, which then in turn controls the flagella’s rotation. This part can be used in the complete recreation of an E. coli’s genetic flagellar pathway. |
| Sources
Motility protein B – Escherichia coli (strain K12). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P0AF06 |
 |
BBa_K842014
T7 RBS cheZ |
Description and Function
T7 serves as a promoter that is stimulated by IPTG. RBS is a sequence in DNA located upstream of the start codon. It affects the rate at which the open reading frame is translated. CheZ is a regulatory protein that constantly dephosphorylates cheY that is attached to a flagellum. The incessant dephosphorylation is what causes bacteria to run, stop, and tumble when in an environment that has a concentration gradient and remain responsive to any changes in chemical concentration. Together, cheZ is induced by the presence of IPTG.
Used for researcher control of signaling mechanisms that influence flagella function.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used with other chemotaxis genes to form a complete regulatory pathway that controls the response system in MCP and the direction of rotation in flagella. |
| Sources
Protein phosphatase CheZ – Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P07800 |
 |
BBa_K842015
T7 RBS cheY |
Description and Function
T7 serves as a promoter that is stimulated by IPTG. RBS is a sequence in DNA located upstream of the start codon. It affects the rate at which the open reading frame is translated. CheY is regulatory protein that reverses the direction of flagella rotation from counter-clockwise to clockwise. When the methyl-accepting chemotaxis protein (MCP) is not bound to any attractant, cheA is activated and told to self-phosphorylate itself by hydrolyzing ATP. Once the cheA is phosphorylated, it transfers its phosphate group to cheY, therefore activating it. CheY then moves to the cytoplasmic side of the flagella apparatus and binds to it. When flagella are not bound to cheY, it rotates in a counter-clockwise direction, which allows the bacteria to move in a “forward” direction. Flagella that are bound to phosphorylated cheY switches its rotation to clockwise, which induces tumbling.
Used for researcher control of signaling mechanisms that influence flagella function.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842015 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used with other chemotaxis genes to form a complete regulatory pathway that controls the response system in MCP and the direction of rotation in flagella. |
| Sources
Chemotaxis protein CheY – Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P0A2D5 |
 |
BBa_K842002
flgJ |
Description and Function
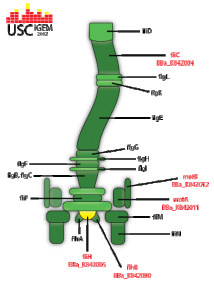
flgJ transcribes a flagellum specific muramidase which hydrolyzes the peptidoglycan layer in the cell membrane. This creates a small hole in the side of the bacterium in which the flagellar rod is formed. It is most commonly believed that flgJ is exported via the flagellum specific exportation system, making it a class 3 flagella gene.
Used for reconstitution of flagella apparatus in E. coli B strains.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used in conjunction with the entire flagella synthesis genetic pathway to reproduce a complete flagella apparatus. Additionally, if this part is used separate from the flagellar genetic pathway and connected to a gene promoter, it will continually react with the cell wall until the peptidoglycan layer of the cell membrane can no longer sustain the structure of the bacterium. |
| Sources
Nambu, T., Minamino, T., Macnab, R., & Kutsukake, K. (1999). Peptidoglycan-Hydrolyzing Activity of the FlgJ Protein, Essential for Flagellar Rod Formation in Salmonella typhimurium. Journal of Bacteriology, 181(5), 1555-1561. Retrieved October 1, 2012, from http://jb.asm.org/content/181/5/1555
Peptidoglycan hydrolase flgJ – Escherichia coli (strain K12). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P75942 |
 |
BBa_K842003
fliA |
Description and Function
fliA is an alternate sigma factor for the class 3 flagella operons. When transcribed, it forms a component of RNA polymerase sigma 28.
Used for reconstitution of flagella apparatus in E. coli B strains.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842002 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used in conjunction with the entire flagella synthesis genetic pathway to reproduce a complete flagella apparatus. |
| Sources
fliA:Quickview – EcoliWiki. (n.d.). EcoliWiki. Retrieved October 1, 2012, from http://ecoliwiki.net/colipedia/index.php/fliA
Ide, N., & Kutsukake, K. (n.d.). Identification of a novel Escherichia coli gene whose e… [Gene. 1997] – PubMed – NCBI. National Center for Biotechnology Information. Retrieved October 1, 2012, from http://www.ncbi.nlm.nih.gov/pubmed/9358034
Claret, L., Miquel, S., Vieille, N., Ryjenkov, D., Gomelsky, M., & Darfeuille-Michaud, A. A. (2007). The Flagellar Sigma Factor FliA Regulates Adhesion and Invasion of Crohn Disease-associated Escherichia coli via a Cyclic Dimeric GMP-dependent Pathway. The Journal Of Biological Chemistry, 282(46), 33275-33283. Retrieved October 1, 2012, from http://www.jbc.org/content/282/46/33275.full.pdf |
 |
BBa_K842005
fliH |
Description and Function
FliH functions as part of the flagella specific export system and is necessary for flagellar growth and assembly. It is believed to be part of a two-component system consisting of fliH and fliL that hydrolyses ATP until construction of the flagella apparatus is completed.
Used for reconstitution of flagella apparatus in E. coli B strains.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842005 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is to be used in conjunction with the entire flagella synthesis genetic pathway to reproduce a complete flagella apparatus. |
| Sources
Minamino, T., & MacNab, R. (n.d.). FliH, a soluble component of the type III flag… [Mol Microbiol. 2000] – PubMed – NCBI. National Center for Biotechnology Information. Retrieved October 1, 2012, from http://www.ncbi.nlm.nih.gov/pubmed/10998179 |
 |
BBa_K842013
fliC-GFP |
Description and Function
When transcribed, RBS-fliC-GFP generates the flagellum protein bonded with a green fluorescent protein. This structure would then be exported to the growing flagella apparatus and used in the construction of the filament. When strongly expressed, this structure would directly result in the flagella filament's containing GFP, allowing for the flagella to be detectable. Unfortunately we were unable to promote the sequence enough to test whether this construct actually functions in this manner.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842013 |
| Cloning and Uses
The fliC gene was cloned via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. Due to the fact that fliC has multiple S and P restriction enzyme sites located within its DNA sequence, it was generated with XbaI sites on the forward and reverse primer. This is so as to allow for the placement of a gene promoter in the E-X sites upstream of the construct in an iGEM biobrick vector. It is important to note though that the current structure consists of the following so as to avoid the accidental loss of a gene during the cloning procedure.
Used for reconstitution of flagella apparatus in E. coli B strains.
EcoRI-XbaI-RBS-fliC-XbaI-GFP-SpeI-PstI |
| Sources
fliC:Quickview – EcoliWiki. (n.d.). EcoliWiki. Retrieved October 1, 2012, from http://ecoliwiki.net/colipedia/index.php/fliC |
 |
BBa_K842000
flhB |
Description and Function
Class 2 flagella operon. When transcribed, flhB is one of three flagella genes responsible for the generation of proteins that export flagellin to the flagella apparatus. The flagellin is then directly used in the synthesis of the structure's basal body. Additionally, flhB directly transcribes the operon flhBAE.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842000 |
| Cloning and Uses
flhB was use generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. It is directly downstream of the class 1 flagella operons flhD and flhC. It also works in conjunction with fliL and fliH to make up the flagellin export system. This part can be used either to generate a flagella in an E. coli strain in which they are not already present, or to promote further formation of pre-existing flagella.
Used for reconstitution of flagella apparatus in E. coli B strains. |
| Sources
Flagellar biosynthetic protein flhB – Escherichia coli (strain K12). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P76299
flhB:Quickview – EcoliWiki. (n.d.). EcoliWiki. Retrieved October 1, 2012, from http://ecoliwiki.net/colipedia/index.php/flhB:Quickview |
 |
BBa_K842001
flhD |
Description and Function
flhD along with flhC form the master regulation operon for the synthesis of flagella in E. coli. They are responsible for the downstream transcription of the class 2 flagella genes. flhD is directly induced by several separate systems; among these is the two-component quorum sensing response system, qseB and qseC.
Used for reconstitution of flagella apparatus in E. coli B strains.
http://partsregistry.org/wiki/index.php?title=Part:BBa_K842001 |
| Cloning and Uses
This part was generated via PCR from the E. coli strain DH5α and cloned into the vector pSB1C3. FlhD is to be used in conjunction with flhC in order to form the transcriptional operon for the production of flagella. This part can be used either to generate a flagella in an E. coli strain in which they are not already present, or to promote further formation of pre-existing flagella. |
| Sources
Flagellar transcriptional regulator FlhD – Escherichia coli (strain K12). (n.d.). UniProt. Retrieved October 1, 2012, from http://www.uniprot.org/uniprot/P0A8S9 |
New Constructs Being Tested:
Because cheY and cheZ have opposing functions, we wanted to test the expression of both in a single E.coli. To ensure equal transcription from this construct, we utilized the commercially availible pCOLA-Duet and pCDF-Duet vectors from Novagen, which have two RBS sites flanking two unique multiple cloning sites, and are all under the control of a single T7 promoter.
These new constructs worked perfectly, and we are currently designing and constructing a similar system in the pSBC1C3 backbone as a new BioBrick.
Although we have not included these in the registry yet, as they have not been fully tested, we have generated the following constructs:
pCOLA-Duet-CheY
pCOLA-Duet-CheZ
pCDF-Duet-CheY
pCDF-Duet-CheZ














 "
"
