Team:Michigan/Project
From 2012.igem.org
(Difference between revisions)
| Line 6: | Line 6: | ||
<h3>Abstract</h3> | <h3>Abstract</h3> | ||
Recombinases can be used to create responsive genetic elements in biological systems. Further, one can build complex control circuits using combinations of invertible DNA sequences. We utilized the unidirectional recombinase HbiF to augment an existing recombinase system in <i>Escherichia coli</i> that relied on the unidirectional recombinase FimE. A burst of low level expression of one recombinase is expected to invert the promoter flanked by the recombinase binding sites IRR and IRL, triggering a switch from strong expression of one to another set of proteins made downstream. Expression of the second recombinase is expected to revert the promoter to its original orientation, triggering the original set of protein expression. The inversion will be sustained across cell divisions with little leaky protein expression and negligible performance degradation after repeated inversions. This is a heritable, binary memory system and can be used as a component in more complex systems. | Recombinases can be used to create responsive genetic elements in biological systems. Further, one can build complex control circuits using combinations of invertible DNA sequences. We utilized the unidirectional recombinase HbiF to augment an existing recombinase system in <i>Escherichia coli</i> that relied on the unidirectional recombinase FimE. A burst of low level expression of one recombinase is expected to invert the promoter flanked by the recombinase binding sites IRR and IRL, triggering a switch from strong expression of one to another set of proteins made downstream. Expression of the second recombinase is expected to revert the promoter to its original orientation, triggering the original set of protein expression. The inversion will be sustained across cell divisions with little leaky protein expression and negligible performance degradation after repeated inversions. This is a heritable, binary memory system and can be used as a component in more complex systems. | ||
| + | |||
| + | <br><br> | ||
<h3>Background</h3> | <h3>Background</h3> | ||
Revision as of 23:56, 3 October 2012
Abstract
Recombinases can be used to create responsive genetic elements in biological systems. Further, one can build complex control circuits using combinations of invertible DNA sequences. We utilized the unidirectional recombinase HbiF to augment an existing recombinase system in Escherichia coli that relied on the unidirectional recombinase FimE. A burst of low level expression of one recombinase is expected to invert the promoter flanked by the recombinase binding sites IRR and IRL, triggering a switch from strong expression of one to another set of proteins made downstream. Expression of the second recombinase is expected to revert the promoter to its original orientation, triggering the original set of protein expression. The inversion will be sustained across cell divisions with little leaky protein expression and negligible performance degradation after repeated inversions. This is a heritable, binary memory system and can be used as a component in more complex systems.Background
fimS or synthetic “flip” sequence
The process of fimbriation in Escherichia coli is regulated by the enzyme-catalyzed inversion of a segment of DNA, often labeled fimS, by a series of tyrosine recombinases of which FimE and FimB are the most known. In the absence of ATP or external prosthetic groups, the fim recombinases introduce strand breaks in repeating regions flanking the fimS regulatory sequence and physically reverse the sequence, activating or repressing the expression of fimbriation proteins based on the orientation of a contained promoter.Inversion catalyzed by FimE was shown to be primarily unidirectional, inverting fimS from the “on” to “off” orientation in regards to fimbriae production. This stands in contrast to FimB, which is bidirectional in inverting fimS, forming an equilibrium between “off” and “on” states.# An inducible genetic circuit utilizing this property was engineered by Ham et al. possessing tightly controllable, strong protein expression with relatively low basal expression and a unique mode of activation, requiring only “pulses” of inducer rather than extended exposure. Team Michigan sought to improve on this idea by utilizing two unidirectional recombinases -- FimE and HbiF.
1.M S McClain et all. “Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli.” J. Bacteriol. September 1991vol. 173 no. 17 5308-5314
Orientation of IRL/IRR Components of fimS dictate enzyme specificity
The fimS region is composed of two inverted repeats, frequently referred to as inverted repeat left (IRL) and inverted repeat right (IRR). The two flank a region of DNA referred to as the inversion region. Each of the inverted repeats are composed of three sections of DNA, an external half-site, an inverted repeat, and a internal half-site. The external half-sites and inverted repeats are stable and identifiers of the IRL and IRR, while the internal half-sites are mobile, switching between the IRL and IRR, dictating recombinase specificity and the orientation of the invertible region. The internal half-sites are inverted along with the inversion region when the region is acted upon by one of the recombinases (Fig 1). (1)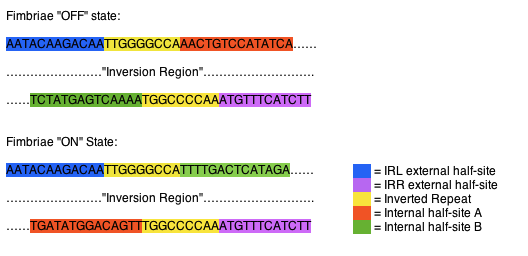
Fig 1. Orientation of fimS inverted repeats. In the “off “state the IRL contains internal half-site A (orange), while the IRR contains internal half-site B (green). When inverted to the “on” state the IRL contains internal half-site B (green), while the IRR contains internal half-site A (orange).
(1) McCuster et al., Molecular Microbiology (2008) 67(1), 171–187
(1) McCuster et al., Molecular Microbiology (2008) 67(1), 171–187
FimE
The FimE enzyme was cloned using the Caltech-made part K137007. A well characterized tyrosine recombinase, the enzyme catalyzes the inversion of a 200-300bp DNA region such that a 3’-facing sequence on the template strand of the region becomes a 3’-facing sequence on the coding strand. This functionality occurs in the lack of general cofactors or ATP, as demonstrated by Ham et al. It its natural state, FimE possesses remarkable catalytic efficiency, with only very low levels capable of flipping regions efficiently.FimE is dependent on properly oriented inverted repeats (IRs) flanking a region of interest for the reaction to occur. Furthermore, it may only invert a fim sequence in the “on” orientation to the “off” orientation.
HbiF
HbiF, a tyrosine recombinase bearing high sequence homology to FimE and other fim recombinases, was discovered in 2006 by Xie et al and also catalyses inversion of the fimS region. Our concept harnesses HbiF to convert the FimE-activatable expression system described by Ham et al. into a bidirectional switch capable of binary states dependent on the presence and expression of either FimE or HbiF recombinases. Team Michigan’s submission of HbiF, BBa_K880000, possesses a protein fusion standard RFC25 suffix to facilitate circuit regulation or protein quantification.In contrast to FimE, HbiF catalyzes the rotation of fimS from the “off” to “on” position in vivo.
Xie et all. “HbiF Regulates Type 1 Fimbriation Independently of FimB and FimE.” Infection and Immunity July 2006 vol. 74 No. 7 4039-4047.
Further Reading
Ham TS, Lee SK, Keasling JD, Arkin AP (2008) Design and Construction of a Double Inversion Recombination Switch for Heritable Sequential Genetic Memory. PLoS ONE 3(7): e2815. doi:10.1371/journal.pone.0002815Ham TS, Lee SK, Keasling JD, Arkin AP (2006) A Tightly Regulated Inducible Expression System Utilizing the fim Inversion Recombination Switch. Biotechnol. Bioeng., Vol. 94(1) doi:10.1002/bit.20916
 "
"

