Team:Wellesley HCI/Notebook/VeronicaNotebook
From 2012.igem.org
(→Jun 21-22: Day 18-19- Continuing with the Crawler) |
|||
| Line 124: | Line 124: | ||
<br> | <br> | ||
| - | == May 29: | + | == May 29: First day of the summer session! == |
Today, we attended the Summer Research Program Orientation - we got to meet our fellow researchers and faculty. Orit presented an introduction about human-computer interaction and the design process we will be following. We received our mentors and were also split into subgroups to start our research. My group - Casey, me and Nicole - is researching semantic search and the importance of it in a synthetic biology setting. | Today, we attended the Summer Research Program Orientation - we got to meet our fellow researchers and faculty. Orit presented an introduction about human-computer interaction and the design process we will be following. We received our mentors and were also split into subgroups to start our research. My group - Casey, me and Nicole - is researching semantic search and the importance of it in a synthetic biology setting. | ||
| - | == May | + | == May 31: Intro to the surface == |
| - | + | I conducted some research, reading up on documents about semantic search and synthetic biology. Later, I learned how to program in C# today through some tutorials on the Microsoft Surface. The tutorials led me through some basic tasks, such as creating an image and allowing it to be resized with a maximum and minimum length and width, grouping multiple objects in categories, and setting an object's initial placement on the surface. For using C# and the Microsoft Surface for the first time, it did not seem too difficult - I am excited to start coding projects on the surface. | |
| - | + | == June 1: More surface tutorials == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == June 1: | + | |
[[Image:Microsoft Design-Day4.png|thumb|Microsoft Design Tutorial]] | [[Image:Microsoft Design-Day4.png|thumb|Microsoft Design Tutorial]] | ||
| Line 142: | Line 138: | ||
I learned about the design principles and guidelines of Microsoft Surface applications. I read about the Windows 8 OS and was introduced to the metro-style apps of the new user interface. Many of the general guidelines were similar to the ones I learned for designing iOS applications, such as focusing on the user and making important objects larger. Other principles were specific to the Windows 8 environment, such as considering how the user navigates the app based on location of controls and gestures. I started the tutorial on the Microsoft surface, but continued it on my computer. Afterwards, I read some more documents and articles about semantic search. | I learned about the design principles and guidelines of Microsoft Surface applications. I read about the Windows 8 OS and was introduced to the metro-style apps of the new user interface. Many of the general guidelines were similar to the ones I learned for designing iOS applications, such as focusing on the user and making important objects larger. Other principles were specific to the Windows 8 environment, such as considering how the user navigates the app based on location of controls and gestures. I started the tutorial on the Microsoft surface, but continued it on my computer. Afterwards, I read some more documents and articles about semantic search. | ||
| - | == June 4: | + | == June 4: Field trip to Microsoft NERD Center! == |
[[Image:NERD_Center-Day5.png|thumb|Microsoft NERD Center in Cambridge, MA]] | [[Image:NERD_Center-Day5.png|thumb|Microsoft NERD Center in Cambridge, MA]] | ||
| Line 150: | Line 146: | ||
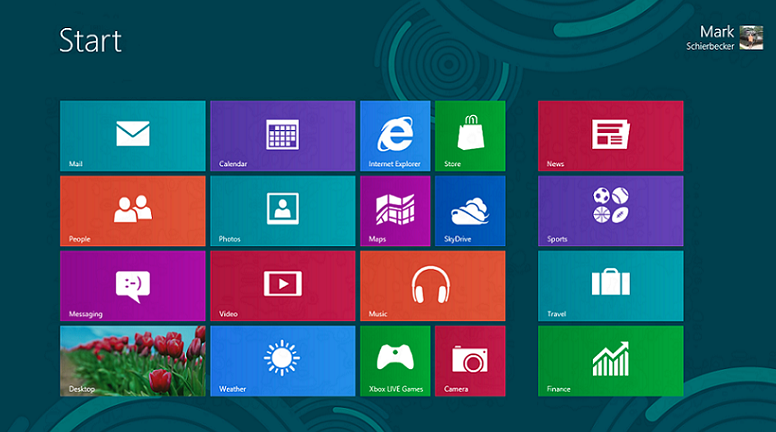
In the morning, I continued my research about semantic search. At lunch, I attended a workshop on Statistics, where we reviewed the concepts of descriptive and inferential statistics. During the afternoon, we all took a field trip to Microsoft NERD Center in Cambridge. The Microsoft building was very interesting - while everything was simple and clean, the colors of the interior made for an environment that was both inspiring and peaceful. After viewing parts of the building, two Microsoft employees sat down with us to chat about Windows 8 and our project. They started with a demo of the new Windows 8 operating system on the new Microsoft tablet. Its first unique feature was the picture password to log in - instead of choosing a numerical password, the user could choose a photo and tap a particular pattern on the photo (i.e. connecting certain faces or going around specific objects) to log in. The next component of the operating system was the main user interface; Windows 8 uses a metro user-interface that displays icons in titled groups. It includes a semantic zoom feature, which allows users to display either the groups, as a big picture, or each group specifically, by pinching the screen. Windows 8 also connects users by enabling users to log in to several social media networks at once and displaying all of the contacts' statuses and photos in one place. Furthermore, the touch-based gestures all follow a different logic - for example, swiping from the right displays a charms menu, swiping from the left switches the application, and swiping down brings up additional menus. I found that the gestures made the tablet fairly difficult to navigate at first, as it took some time and practice to get used to all the various gestures. After watching the Windows 8 demo, we learned about developing metro-style apps through Visual Studio 11 and Blend for Visual Studio. It was similar to Xcode in the way that the developer could drag objects into the view and write code for more detailed functionality. Ultimately, the most important thing I learned was the semantic zoom feature of the Windows 8 OS - since my team is researching semantic search and semantic zoom, it was encouraging to see how easily semantic zoom could be integrated into the apps. Semantic zoom would allow the users to view the provided information at a glance without scrolling through all of the information. Overall, it was a very productive day. | In the morning, I continued my research about semantic search. At lunch, I attended a workshop on Statistics, where we reviewed the concepts of descriptive and inferential statistics. During the afternoon, we all took a field trip to Microsoft NERD Center in Cambridge. The Microsoft building was very interesting - while everything was simple and clean, the colors of the interior made for an environment that was both inspiring and peaceful. After viewing parts of the building, two Microsoft employees sat down with us to chat about Windows 8 and our project. They started with a demo of the new Windows 8 operating system on the new Microsoft tablet. Its first unique feature was the picture password to log in - instead of choosing a numerical password, the user could choose a photo and tap a particular pattern on the photo (i.e. connecting certain faces or going around specific objects) to log in. The next component of the operating system was the main user interface; Windows 8 uses a metro user-interface that displays icons in titled groups. It includes a semantic zoom feature, which allows users to display either the groups, as a big picture, or each group specifically, by pinching the screen. Windows 8 also connects users by enabling users to log in to several social media networks at once and displaying all of the contacts' statuses and photos in one place. Furthermore, the touch-based gestures all follow a different logic - for example, swiping from the right displays a charms menu, swiping from the left switches the application, and swiping down brings up additional menus. I found that the gestures made the tablet fairly difficult to navigate at first, as it took some time and practice to get used to all the various gestures. After watching the Windows 8 demo, we learned about developing metro-style apps through Visual Studio 11 and Blend for Visual Studio. It was similar to Xcode in the way that the developer could drag objects into the view and write code for more detailed functionality. Ultimately, the most important thing I learned was the semantic zoom feature of the Windows 8 OS - since my team is researching semantic search and semantic zoom, it was encouraging to see how easily semantic zoom could be integrated into the apps. Semantic zoom would allow the users to view the provided information at a glance without scrolling through all of the information. Overall, it was a very productive day. | ||
| - | == Jun 5: | + | == Jun 5: A day in the wet lab == |
[[Image:Wetlab MIT-Day6.jpg|thumb|Watching Professor Kuldell's presentation]] | [[Image:Wetlab MIT-Day6.jpg|thumb|Watching Professor Kuldell's presentation]] | ||
| Line 160: | Line 156: | ||
We spent the entire day at MIT with Professor Natalie Kuldell. In the morning, she presented several lectures. She introduced the field of synthetic biology and explained the typical process of synthesis, abstraction, and standardization. We learned about the more technical side of synthetic biology - how researchers find the section of the gene they want, how the cell duplicates genes, etc. Professor Kuldell then discussed "Eau d'coli," an iGEM submission from MIT in 2006. It was very interesting to learn about their project, as it is a real application of synthetic biology that our software project aims to improve. MIT's goal was to genetically engineer E.coli cells to smell like banana and wintergreen. We covered their entire process, as well as that of the cells and cell parts. In addition, we watched a video of MIT's presentation at iGEM to fully understand the challenges they came across. As I learned, I connected their process with our semantic search project, and wondered a couple things - how did they search the smells and narrow them down to only banana and wintergreen? How did they know what promoter to use? Was it written somewhere that indole was what produced the natural smell of E.coli? After Professor Kuldell's lectures, we went through a lab safety lecture and then headed to the lab to conduct some experiments of our own. The first lab, titled "What a Color World," was related to the MIT iGEM's "Eau d'coli" project. We prepared 2 different E.coli strains for transformation and then transformed them with purple-color generator and green-color generator. In the second lab, titled "iTunes device," we examined promoter and RBS (ribosome-binding site) combinations to optimize beta-galactosidase output. We used test tubes, pipets, petri dishes, and a spectrophotometer to determine which combination was the best. I was shocked by how precise the measurements had to be, the length of time it took to see any results in experiments, and how much work had to be done to prove one small thing. Our day ended with an overview of the day and our impressions of synthetic biology. | We spent the entire day at MIT with Professor Natalie Kuldell. In the morning, she presented several lectures. She introduced the field of synthetic biology and explained the typical process of synthesis, abstraction, and standardization. We learned about the more technical side of synthetic biology - how researchers find the section of the gene they want, how the cell duplicates genes, etc. Professor Kuldell then discussed "Eau d'coli," an iGEM submission from MIT in 2006. It was very interesting to learn about their project, as it is a real application of synthetic biology that our software project aims to improve. MIT's goal was to genetically engineer E.coli cells to smell like banana and wintergreen. We covered their entire process, as well as that of the cells and cell parts. In addition, we watched a video of MIT's presentation at iGEM to fully understand the challenges they came across. As I learned, I connected their process with our semantic search project, and wondered a couple things - how did they search the smells and narrow them down to only banana and wintergreen? How did they know what promoter to use? Was it written somewhere that indole was what produced the natural smell of E.coli? After Professor Kuldell's lectures, we went through a lab safety lecture and then headed to the lab to conduct some experiments of our own. The first lab, titled "What a Color World," was related to the MIT iGEM's "Eau d'coli" project. We prepared 2 different E.coli strains for transformation and then transformed them with purple-color generator and green-color generator. In the second lab, titled "iTunes device," we examined promoter and RBS (ribosome-binding site) combinations to optimize beta-galactosidase output. We used test tubes, pipets, petri dishes, and a spectrophotometer to determine which combination was the best. I was shocked by how precise the measurements had to be, the length of time it took to see any results in experiments, and how much work had to be done to prove one small thing. Our day ended with an overview of the day and our impressions of synthetic biology. | ||
| - | == Jun 6: | + | == Jun 6: Continuing research == |
We spent the day conducting more research and preparing for our presentations. My team discussed the possibilities of using semantic search in a synthetic biology environment. We examined several semantic search engines with APIs we could potentially implement. In addition, we brainstormed advantages of using semantic search and semantic zoom. | We spent the day conducting more research and preparing for our presentations. My team discussed the possibilities of using semantic search in a synthetic biology environment. We examined several semantic search engines with APIs we could potentially implement. In addition, we brainstormed advantages of using semantic search and semantic zoom. | ||
| - | == Jun 7: | + | == Jun 7: More research == |
[[Image:VL Birthday-Day8.jpg|thumb|Celebrating my birthday in the lab!]] | [[Image:VL Birthday-Day8.jpg|thumb|Celebrating my birthday in the lab!]] | ||
| Line 170: | Line 166: | ||
Today's my birthday! The lab surprised me with a cake, lots of fruit, and ice cream at lunch...probably the best study break ever. After researching in the morning, we met with Orit and Consuelo in the afternoon. They gave us several helpful suggestions - they encouraged us to research more about how semantic search and semantic zoom can be implemented in a multitouch environment, and how it is relevant in human-computer interaction. They also asked us to check out specific API's to determine the feasibility of semantic search in our application. In addition, they gave us more direction with the research, suggesting that we look up abstracts on ACM's (Association for Computing Machinery) Digital Library. | Today's my birthday! The lab surprised me with a cake, lots of fruit, and ice cream at lunch...probably the best study break ever. After researching in the morning, we met with Orit and Consuelo in the afternoon. They gave us several helpful suggestions - they encouraged us to research more about how semantic search and semantic zoom can be implemented in a multitouch environment, and how it is relevant in human-computer interaction. They also asked us to check out specific API's to determine the feasibility of semantic search in our application. In addition, they gave us more direction with the research, suggesting that we look up abstracts on ACM's (Association for Computing Machinery) Digital Library. | ||
| - | == Jun 8: | + | == Jun 8: Even more research == |
After exploring some of Orit and Consuelo's suggestions in the morning, we met with Eni, another computer science professor at Wellesley who is more familiar with the semantic web. She gave us many tips and ideas to explore. She explained RDF and OWL, and suggested an article from the American Scientific Journal about the origins of the word "semantic." She also encouraged us to check out Google's Knowledge Graph, which is essentially the same thing as a semantic search engine. Eni referred us to the International Conference on the Semantic Web, and then gave us short summaries of Apache Jena and Apache Lucene to help us with possibilities of implementing semantic search and semantic zoom. Overall, it was a very helpful meeting - we spent the remainder of the afternoon researching her suggestions. | After exploring some of Orit and Consuelo's suggestions in the morning, we met with Eni, another computer science professor at Wellesley who is more familiar with the semantic web. She gave us many tips and ideas to explore. She explained RDF and OWL, and suggested an article from the American Scientific Journal about the origins of the word "semantic." She also encouraged us to check out Google's Knowledge Graph, which is essentially the same thing as a semantic search engine. Eni referred us to the International Conference on the Semantic Web, and then gave us short summaries of Apache Jena and Apache Lucene to help us with possibilities of implementing semantic search and semantic zoom. Overall, it was a very helpful meeting - we spent the remainder of the afternoon researching her suggestions. | ||
| - | == Jun 11: | + | == Jun 11: Presenting the research == |
[[Image:Brainstorming2-Day10.jpg|thumb|Starting the brainstorming process]] | [[Image:Brainstorming2-Day10.jpg|thumb|Starting the brainstorming process]] | ||
| Line 182: | Line 178: | ||
Today we wrapped up the research on semantic search. We spent half of the morning putting our presentation together, and we spent the other half watching Madeline Albright and Hillary Clinton speak at the Women in Public Service Institute opening ceremony. We began our presentations right after lunch. Each group presented their topic for about 15-30 minutes, and then the entire lab group discussed the topic and brainstormed potential challenges, features and methods of implementation. After each group finished, our lab team split into two groups, each of which came up with 5 different project ideas. My group had a variety of ideas - one was a "bubble-themed" way of organizing the menu and data on a tabletop surface. Another was using the paper-lens feature to display different characteristics of a gene through a projection. The day enabled me to learn about other topics in Human Computer Interaction and to brainstorm ambitiously about potential projects. | Today we wrapped up the research on semantic search. We spent half of the morning putting our presentation together, and we spent the other half watching Madeline Albright and Hillary Clinton speak at the Women in Public Service Institute opening ceremony. We began our presentations right after lunch. Each group presented their topic for about 15-30 minutes, and then the entire lab group discussed the topic and brainstormed potential challenges, features and methods of implementation. After each group finished, our lab team split into two groups, each of which came up with 5 different project ideas. My group had a variety of ideas - one was a "bubble-themed" way of organizing the menu and data on a tabletop surface. Another was using the paper-lens feature to display different characteristics of a gene through a projection. The day enabled me to learn about other topics in Human Computer Interaction and to brainstorm ambitiously about potential projects. | ||
| - | == Jun 12: | + | == Jun 12: Another day in the wet lab == |
[[Image:BUDay11.jpg|thumb|Learning at BU]] | [[Image:BUDay11.jpg|thumb|Learning at BU]] | ||
| Line 188: | Line 184: | ||
We spent the entire day at Boston University's Photonics Building with their iGEM wet lab team. We began the morning with an introduction to synthetic biology and biology basics: we learned the definition of synthetic biology - as quoted from Ahmad Khalil and James Collins, two well-known researchers in the field, "Synthetic biology is bringing together engineers and biologists to design and build novel bimolecular components, networks and pathways, and to use these constructs to rewire and reprogram organisms." We then went on to cover the parts of a transcriptional unit, including the definitions of a promoter, a ribosome binding site, a gene and a terminator. In addition, we reviewed plasmids and bacterial transformation. Finally, we learned about the methods that the BU wet lab team is using; knowledge of their experimental process enables us to better understand the needs of our potential users. We also shared our researched features and potential projects. Overall, it was a valuable experience to learn about a real wet lab team's process and to share our ideas. | We spent the entire day at Boston University's Photonics Building with their iGEM wet lab team. We began the morning with an introduction to synthetic biology and biology basics: we learned the definition of synthetic biology - as quoted from Ahmad Khalil and James Collins, two well-known researchers in the field, "Synthetic biology is bringing together engineers and biologists to design and build novel bimolecular components, networks and pathways, and to use these constructs to rewire and reprogram organisms." We then went on to cover the parts of a transcriptional unit, including the definitions of a promoter, a ribosome binding site, a gene and a terminator. In addition, we reviewed plasmids and bacterial transformation. Finally, we learned about the methods that the BU wet lab team is using; knowledge of their experimental process enables us to better understand the needs of our potential users. We also shared our researched features and potential projects. Overall, it was a valuable experience to learn about a real wet lab team's process and to share our ideas. | ||
| - | == Jun 13: | + | == Jun 13: Demoing and Brainstorming! == |
[[Image:Demoing-Day12.jpg|thumb|Demoing the Beast to Agilent rep]] | [[Image:Demoing-Day12.jpg|thumb|Demoing the Beast to Agilent rep]] | ||
| Line 196: | Line 192: | ||
As a continuation of yesterday's brainstorming, the Boston University iGEM team came to Wellesley today. Before they arrived, we gave a demo of our Beast surface and its applications to a representative from Agilent. He gave us some thoughtful feedback. In the afternoon, we all gathered in a room and covered the walls with ideas. We had four different categories - the Lab Organization Tool (originally the eLab Notebook), the Beast (both micro and macro features), Semantic Search, and Art. For the first hour, everyone wandered around the room, exploring the ideas and notes already written and then adding their own. Then, we sat together as a group and discussed each topic. We came up with several great ideas for each topic, and we have our work clearly cut out for us. | As a continuation of yesterday's brainstorming, the Boston University iGEM team came to Wellesley today. Before they arrived, we gave a demo of our Beast surface and its applications to a representative from Agilent. He gave us some thoughtful feedback. In the afternoon, we all gathered in a room and covered the walls with ideas. We had four different categories - the Lab Organization Tool (originally the eLab Notebook), the Beast (both micro and macro features), Semantic Search, and Art. For the first hour, everyone wandered around the room, exploring the ideas and notes already written and then adding their own. Then, we sat together as a group and discussed each topic. We came up with several great ideas for each topic, and we have our work clearly cut out for us. | ||
| - | == Jun 14-15: | + | == Jun 14-15: More brainstorming == |
[[Image:More Brainstorming-Day13.jpg|thumb|More brainstorming of project ideas]] | [[Image:More Brainstorming-Day13.jpg|thumb|More brainstorming of project ideas]] | ||
| Line 202: | Line 198: | ||
Unfortunately, there was a personal emergency and I was unable to show up in the lab these 2 days. From my understanding, the lab continued researching and brainstorming. My team aimed to look at the registry and attempt to convert it to RDF to investigate how easily semantic search could be implemented. | Unfortunately, there was a personal emergency and I was unable to show up in the lab these 2 days. From my understanding, the lab continued researching and brainstorming. My team aimed to look at the registry and attempt to convert it to RDF to investigate how easily semantic search could be implemented. | ||
| - | == Jun 18: | + | == Jun 18: Brainstorming and Researching == |
Today, I spent the morning catching up on all the brainstorming I missed. In the afternoon, I researched previous iGEM teams on the iGEM archives to see if any teams had already converted the iGEM registry into RDF format. I found a couple teams who had worked with the registry, but none of them converted it to RDF. At the end of the day, we decided to approach the problem differently. | Today, I spent the morning catching up on all the brainstorming I missed. In the afternoon, I researched previous iGEM teams on the iGEM archives to see if any teams had already converted the iGEM registry into RDF format. I found a couple teams who had worked with the registry, but none of them converted it to RDF. At the end of the day, we decided to approach the problem differently. | ||
| - | == Jun 19: | + | == Jun 19: Using Google Custom Search == |
We examined other ways access data from the Parts Registry and the iGEM registry. First, we looked into using a web crawler. However, we decided to try mining data from Google Custom Search first. We figured out how to use the Google Custom Search results through Visual Studio and have the results display in a data grid. We had a team meeting with Orit in the afternoon to discuss the MoClo Planner and the work we had done so far - she was happy we had figured out how to implement Google Custom Search. | We examined other ways access data from the Parts Registry and the iGEM registry. First, we looked into using a web crawler. However, we decided to try mining data from Google Custom Search first. We figured out how to use the Google Custom Search results through Visual Studio and have the results display in a data grid. We had a team meeting with Orit in the afternoon to discuss the MoClo Planner and the work we had done so far - she was happy we had figured out how to implement Google Custom Search. | ||
| - | == Jun 20: | + | == Jun 20: Creating a Web Crawler == |
As we examined our work from yesterday, we realized that our data grid of the Google Custom Search results could only display 10 results, and those 10 results did not match those that came up online. Again, we had to abandon that approach. We resorted to creating a crawler, using a previous PubMed crawler as a template. Our first job was to customize it to pull data from the Parts Registry. | As we examined our work from yesterday, we realized that our data grid of the Google Custom Search results could only display 10 results, and those 10 results did not match those that came up online. Again, we had to abandon that approach. We resorted to creating a crawler, using a previous PubMed crawler as a template. Our first job was to customize it to pull data from the Parts Registry. | ||
| - | == Jun 21-22: | + | == Jun 21-22: Continuing with the Crawler == |
Nicole and I continued to work on the back-end - we created two files: one to access the data on the Parts Registry and one to grab specific data to put in each part's data sheet. We worked with a few other team members to come up with a comprehensive data sheet template. | Nicole and I continued to work on the back-end - we created two files: one to access the data on the Parts Registry and one to grab specific data to put in each part's data sheet. We worked with a few other team members to come up with a comprehensive data sheet template. | ||
| + | |||
| + | == Jun 25: Still working on the Crawler == | ||
| + | |||
| + | Today, we made some more progress on the web crawler - we got RegList to extract data from the Parts Registry. We also used the tentative data sheet template to do some basic coding on RegDataSheet. Nicole and I are getting better at pair programming. | ||
| + | |||
| + | == Jun 26: Almost finished with the Crawler == | ||
| + | |||
| + | We're getting closer to finishing the Parts Registry web crawler. We worked on it all day, and by the end of the day, we were able to have the console print out a tentative "data sheet," complete with the name of the part, its link, its type, and its regulation. It also showed some basic information, including the part's SBOL image, its availability and usefulness, its sequence, its length and number of twins. Finally, it could display some protocol information as well as references, including the authors of the part, the group and the date. | ||
<!--notebook ends here--> | <!--notebook ends here--> | ||
Revision as of 03:15, 6 July 2012
Veronica's Notebook
May 29: First day of the summer session!
Today, we attended the Summer Research Program Orientation - we got to meet our fellow researchers and faculty. Orit presented an introduction about human-computer interaction and the design process we will be following. We received our mentors and were also split into subgroups to start our research. My group - Casey, me and Nicole - is researching semantic search and the importance of it in a synthetic biology setting.
May 31: Intro to the surface
I conducted some research, reading up on documents about semantic search and synthetic biology. Later, I learned how to program in C# today through some tutorials on the Microsoft Surface. The tutorials led me through some basic tasks, such as creating an image and allowing it to be resized with a maximum and minimum length and width, grouping multiple objects in categories, and setting an object's initial placement on the surface. For using C# and the Microsoft Surface for the first time, it did not seem too difficult - I am excited to start coding projects on the surface.
June 1: More surface tutorials
I learned about the design principles and guidelines of Microsoft Surface applications. I read about the Windows 8 OS and was introduced to the metro-style apps of the new user interface. Many of the general guidelines were similar to the ones I learned for designing iOS applications, such as focusing on the user and making important objects larger. Other principles were specific to the Windows 8 environment, such as considering how the user navigates the app based on location of controls and gestures. I started the tutorial on the Microsoft surface, but continued it on my computer. Afterwards, I read some more documents and articles about semantic search.
June 4: Field trip to Microsoft NERD Center!
In the morning, I continued my research about semantic search. At lunch, I attended a workshop on Statistics, where we reviewed the concepts of descriptive and inferential statistics. During the afternoon, we all took a field trip to Microsoft NERD Center in Cambridge. The Microsoft building was very interesting - while everything was simple and clean, the colors of the interior made for an environment that was both inspiring and peaceful. After viewing parts of the building, two Microsoft employees sat down with us to chat about Windows 8 and our project. They started with a demo of the new Windows 8 operating system on the new Microsoft tablet. Its first unique feature was the picture password to log in - instead of choosing a numerical password, the user could choose a photo and tap a particular pattern on the photo (i.e. connecting certain faces or going around specific objects) to log in. The next component of the operating system was the main user interface; Windows 8 uses a metro user-interface that displays icons in titled groups. It includes a semantic zoom feature, which allows users to display either the groups, as a big picture, or each group specifically, by pinching the screen. Windows 8 also connects users by enabling users to log in to several social media networks at once and displaying all of the contacts' statuses and photos in one place. Furthermore, the touch-based gestures all follow a different logic - for example, swiping from the right displays a charms menu, swiping from the left switches the application, and swiping down brings up additional menus. I found that the gestures made the tablet fairly difficult to navigate at first, as it took some time and practice to get used to all the various gestures. After watching the Windows 8 demo, we learned about developing metro-style apps through Visual Studio 11 and Blend for Visual Studio. It was similar to Xcode in the way that the developer could drag objects into the view and write code for more detailed functionality. Ultimately, the most important thing I learned was the semantic zoom feature of the Windows 8 OS - since my team is researching semantic search and semantic zoom, it was encouraging to see how easily semantic zoom could be integrated into the apps. Semantic zoom would allow the users to view the provided information at a glance without scrolling through all of the information. Overall, it was a very productive day.
Jun 5: A day in the wet lab
We spent the entire day at MIT with Professor Natalie Kuldell. In the morning, she presented several lectures. She introduced the field of synthetic biology and explained the typical process of synthesis, abstraction, and standardization. We learned about the more technical side of synthetic biology - how researchers find the section of the gene they want, how the cell duplicates genes, etc. Professor Kuldell then discussed "Eau d'coli," an iGEM submission from MIT in 2006. It was very interesting to learn about their project, as it is a real application of synthetic biology that our software project aims to improve. MIT's goal was to genetically engineer E.coli cells to smell like banana and wintergreen. We covered their entire process, as well as that of the cells and cell parts. In addition, we watched a video of MIT's presentation at iGEM to fully understand the challenges they came across. As I learned, I connected their process with our semantic search project, and wondered a couple things - how did they search the smells and narrow them down to only banana and wintergreen? How did they know what promoter to use? Was it written somewhere that indole was what produced the natural smell of E.coli? After Professor Kuldell's lectures, we went through a lab safety lecture and then headed to the lab to conduct some experiments of our own. The first lab, titled "What a Color World," was related to the MIT iGEM's "Eau d'coli" project. We prepared 2 different E.coli strains for transformation and then transformed them with purple-color generator and green-color generator. In the second lab, titled "iTunes device," we examined promoter and RBS (ribosome-binding site) combinations to optimize beta-galactosidase output. We used test tubes, pipets, petri dishes, and a spectrophotometer to determine which combination was the best. I was shocked by how precise the measurements had to be, the length of time it took to see any results in experiments, and how much work had to be done to prove one small thing. Our day ended with an overview of the day and our impressions of synthetic biology.
Jun 6: Continuing research
We spent the day conducting more research and preparing for our presentations. My team discussed the possibilities of using semantic search in a synthetic biology environment. We examined several semantic search engines with APIs we could potentially implement. In addition, we brainstormed advantages of using semantic search and semantic zoom.
Jun 7: More research
Today's my birthday! The lab surprised me with a cake, lots of fruit, and ice cream at lunch...probably the best study break ever. After researching in the morning, we met with Orit and Consuelo in the afternoon. They gave us several helpful suggestions - they encouraged us to research more about how semantic search and semantic zoom can be implemented in a multitouch environment, and how it is relevant in human-computer interaction. They also asked us to check out specific API's to determine the feasibility of semantic search in our application. In addition, they gave us more direction with the research, suggesting that we look up abstracts on ACM's (Association for Computing Machinery) Digital Library.
Jun 8: Even more research
After exploring some of Orit and Consuelo's suggestions in the morning, we met with Eni, another computer science professor at Wellesley who is more familiar with the semantic web. She gave us many tips and ideas to explore. She explained RDF and OWL, and suggested an article from the American Scientific Journal about the origins of the word "semantic." She also encouraged us to check out Google's Knowledge Graph, which is essentially the same thing as a semantic search engine. Eni referred us to the International Conference on the Semantic Web, and then gave us short summaries of Apache Jena and Apache Lucene to help us with possibilities of implementing semantic search and semantic zoom. Overall, it was a very helpful meeting - we spent the remainder of the afternoon researching her suggestions.
Jun 11: Presenting the research
Today we wrapped up the research on semantic search. We spent half of the morning putting our presentation together, and we spent the other half watching Madeline Albright and Hillary Clinton speak at the Women in Public Service Institute opening ceremony. We began our presentations right after lunch. Each group presented their topic for about 15-30 minutes, and then the entire lab group discussed the topic and brainstormed potential challenges, features and methods of implementation. After each group finished, our lab team split into two groups, each of which came up with 5 different project ideas. My group had a variety of ideas - one was a "bubble-themed" way of organizing the menu and data on a tabletop surface. Another was using the paper-lens feature to display different characteristics of a gene through a projection. The day enabled me to learn about other topics in Human Computer Interaction and to brainstorm ambitiously about potential projects.
Jun 12: Another day in the wet lab
We spent the entire day at Boston University's Photonics Building with their iGEM wet lab team. We began the morning with an introduction to synthetic biology and biology basics: we learned the definition of synthetic biology - as quoted from Ahmad Khalil and James Collins, two well-known researchers in the field, "Synthetic biology is bringing together engineers and biologists to design and build novel bimolecular components, networks and pathways, and to use these constructs to rewire and reprogram organisms." We then went on to cover the parts of a transcriptional unit, including the definitions of a promoter, a ribosome binding site, a gene and a terminator. In addition, we reviewed plasmids and bacterial transformation. Finally, we learned about the methods that the BU wet lab team is using; knowledge of their experimental process enables us to better understand the needs of our potential users. We also shared our researched features and potential projects. Overall, it was a valuable experience to learn about a real wet lab team's process and to share our ideas.
Jun 13: Demoing and Brainstorming!
As a continuation of yesterday's brainstorming, the Boston University iGEM team came to Wellesley today. Before they arrived, we gave a demo of our Beast surface and its applications to a representative from Agilent. He gave us some thoughtful feedback. In the afternoon, we all gathered in a room and covered the walls with ideas. We had four different categories - the Lab Organization Tool (originally the eLab Notebook), the Beast (both micro and macro features), Semantic Search, and Art. For the first hour, everyone wandered around the room, exploring the ideas and notes already written and then adding their own. Then, we sat together as a group and discussed each topic. We came up with several great ideas for each topic, and we have our work clearly cut out for us.
Jun 14-15: More brainstorming
Unfortunately, there was a personal emergency and I was unable to show up in the lab these 2 days. From my understanding, the lab continued researching and brainstorming. My team aimed to look at the registry and attempt to convert it to RDF to investigate how easily semantic search could be implemented.
Jun 18: Brainstorming and Researching
Today, I spent the morning catching up on all the brainstorming I missed. In the afternoon, I researched previous iGEM teams on the iGEM archives to see if any teams had already converted the iGEM registry into RDF format. I found a couple teams who had worked with the registry, but none of them converted it to RDF. At the end of the day, we decided to approach the problem differently.
Jun 19: Using Google Custom Search
We examined other ways access data from the Parts Registry and the iGEM registry. First, we looked into using a web crawler. However, we decided to try mining data from Google Custom Search first. We figured out how to use the Google Custom Search results through Visual Studio and have the results display in a data grid. We had a team meeting with Orit in the afternoon to discuss the MoClo Planner and the work we had done so far - she was happy we had figured out how to implement Google Custom Search.
Jun 20: Creating a Web Crawler
As we examined our work from yesterday, we realized that our data grid of the Google Custom Search results could only display 10 results, and those 10 results did not match those that came up online. Again, we had to abandon that approach. We resorted to creating a crawler, using a previous PubMed crawler as a template. Our first job was to customize it to pull data from the Parts Registry.
Jun 21-22: Continuing with the Crawler
Nicole and I continued to work on the back-end - we created two files: one to access the data on the Parts Registry and one to grab specific data to put in each part's data sheet. We worked with a few other team members to come up with a comprehensive data sheet template.
Jun 25: Still working on the Crawler
Today, we made some more progress on the web crawler - we got RegList to extract data from the Parts Registry. We also used the tentative data sheet template to do some basic coding on RegDataSheet. Nicole and I are getting better at pair programming.
Jun 26: Almost finished with the Crawler
We're getting closer to finishing the Parts Registry web crawler. We worked on it all day, and by the end of the day, we were able to have the console print out a tentative "data sheet," complete with the name of the part, its link, its type, and its regulation. It also showed some basic information, including the part's SBOL image, its availability and usefulness, its sequence, its length and number of twins. Finally, it could display some protocol information as well as references, including the authors of the part, the group and the date.
 "
"